Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN4483-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN4483-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 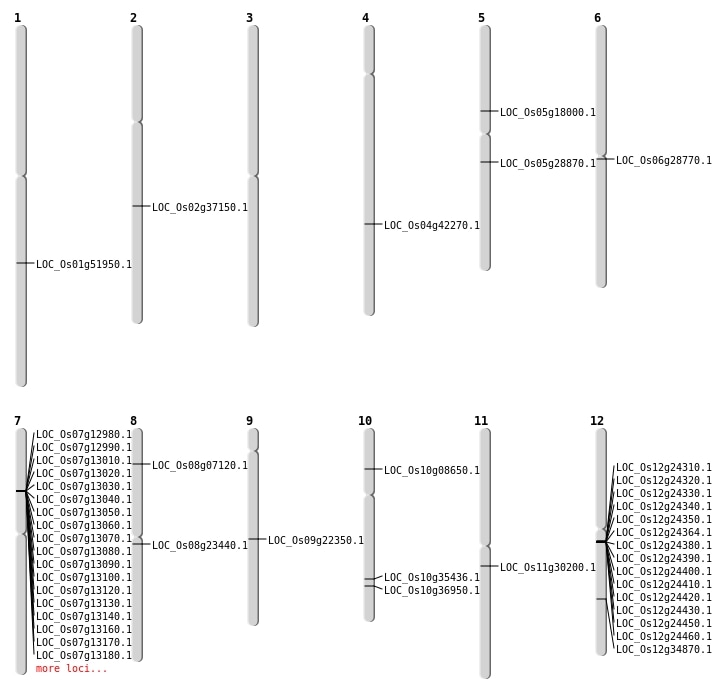
|
| Variant Information |
|---|
Single base substitutions: 34
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 1246612 | G-T | HET | INTERGENIC | |
| SBS | Chr1 | 25984481 | C-T | HET | INTERGENIC | |
| SBS | Chr1 | 40112599 | G-A | HET | INTERGENIC | |
| SBS | Chr10 | 11163229 | T-C | HOMO | INTERGENIC | |
| SBS | Chr10 | 4688111 | T-C | HET | NON_SYNONYMOUS_CODING | LOC_Os10g08650 |
| SBS | Chr10 | 5295578 | C-T | HOMO | INTRON | |
| SBS | Chr10 | 5847239 | G-A | HOMO | INTERGENIC | |
| SBS | Chr11 | 11680983 | C-G | HET | INTERGENIC | |
| SBS | Chr11 | 11680984 | G-T | HET | INTERGENIC | |
| SBS | Chr11 | 1480409 | G-T | HET | INTERGENIC | |
| SBS | Chr11 | 23057673 | A-C | HET | INTERGENIC | |
| SBS | Chr12 | 21228946 | A-T | HOMO | NON_SYNONYMOUS_CODING | LOC_Os12g34870 |
| SBS | Chr12 | 21320543 | T-A | HET | INTRON | |
| SBS | Chr2 | 18292238 | G-A | HET | INTRON | |
| SBS | Chr2 | 29781648 | C-A | HOMO | INTERGENIC | |
| SBS | Chr3 | 14307349 | A-T | HET | INTERGENIC | |
| SBS | Chr3 | 18832171 | C-A | HET | INTERGENIC | |
| SBS | Chr3 | 19848678 | C-T | HET | INTERGENIC | |
| SBS | Chr3 | 4927962 | G-A | HOMO | INTERGENIC | |
| SBS | Chr4 | 10799937 | G-A | HOMO | INTERGENIC | |
| SBS | Chr4 | 20974308 | A-G | HET | INTERGENIC | |
| SBS | Chr4 | 21656545 | A-C | HOMO | INTERGENIC | |
| SBS | Chr4 | 27645692 | A-G | HET | UTR_5_PRIME | |
| SBS | Chr4 | 28347145 | C-T | HET | INTERGENIC | |
| SBS | Chr4 | 6358827 | C-T | HOMO | SYNONYMOUS_CODING | |
| SBS | Chr5 | 1415782 | T-C | HET | INTERGENIC | |
| SBS | Chr5 | 22799397 | T-C | HET | INTERGENIC | |
| SBS | Chr5 | 5764703 | C-G | HET | INTRON | |
| SBS | Chr6 | 27564691 | C-T | HET | INTERGENIC | |
| SBS | Chr7 | 19379639 | A-T | HOMO | INTRON | |
| SBS | Chr8 | 24287020 | G-A | HOMO | INTRON | |
| SBS | Chr8 | 26057396 | G-A | HET | INTERGENIC | |
| SBS | Chr9 | 10822406 | G-T | HET | INTERGENIC | |
| SBS | Chr9 | 13507428 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os09g22350 |
Deletions: 32
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr11 | 4209219 | 4209220 | 1 | |
| Deletion | Chr5 | 5134396 | 5134486 | 90 | |
| Deletion | Chr4 | 5222910 | 5222912 | 2 | |
| Deletion | Chr12 | 6673814 | 6673815 | 1 | |
| Deletion | Chr10 | 6988351 | 6988352 | 1 | |
| Deletion | Chr7 | 7450001 | 7648000 | 198000 | 31 |
| Deletion | Chr2 | 7498119 | 7498125 | 6 | |
| Deletion | Chr5 | 10352396 | 10352397 | 1 | LOC_Os05g18000 |
| Deletion | Chr2 | 11311985 | 11311986 | 1 | |
| Deletion | Chr3 | 12280985 | 12280994 | 9 | |
| Deletion | Chr12 | 13090058 | 13090060 | 2 | |
| Deletion | Chr12 | 13646740 | 13646741 | 1 | |
| Deletion | Chr12 | 13868001 | 13974000 | 106000 | 14 |
| Deletion | Chr3 | 14105074 | 14105128 | 54 | |
| Deletion | Chr8 | 14221169 | 14221174 | 5 | |
| Deletion | Chr12 | 14685624 | 14685625 | 1 | |
| Deletion | Chr12 | 14913406 | 14913407 | 1 | |
| Deletion | Chr5 | 16931572 | 16931573 | 1 | LOC_Os05g28870 |
| Deletion | Chr3 | 18095456 | 18095457 | 1 | |
| Deletion | Chr11 | 19855181 | 19855182 | 1 | |
| Deletion | Chr7 | 20530580 | 20530594 | 14 | |
| Deletion | Chr7 | 22152698 | 22152718 | 20 | |
| Deletion | Chr2 | 22879139 | 22879141 | 2 | |
| Deletion | Chr6 | 23305343 | 23305344 | 1 | |
| Deletion | Chr4 | 25008434 | 25008435 | 1 | LOC_Os04g42270 |
| Deletion | Chr11 | 27177320 | 27177326 | 6 | |
| Deletion | Chr5 | 27811800 | 27811802 | 2 | |
| Deletion | Chr5 | 28856069 | 28856070 | 1 | |
| Deletion | Chr1 | 29861491 | 29861494 | 3 | LOC_Os01g51950 |
| Deletion | Chr1 | 29866893 | 29866895 | 2 | |
| Deletion | Chr1 | 30664192 | 30664193 | 1 | |
| Deletion | Chr3 | 32353910 | 32353918 | 8 |
Insertions: 16
Inversions: 6
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr3 | 2349568 | 2349797 | |
| Inversion | Chr8 | 3986102 | 3986364 | LOC_Os08g07120 |
| Inversion | Chr9 | 16673210 | 16673426 | |
| Inversion | Chr11 | 17017084 | 17559487 | LOC_Os11g30200 |
| Inversion | Chr11 | 17017098 | 17559537 | LOC_Os11g30200 |
| Inversion | Chr7 | 18340684 | 19328207 | LOC_Os07g30980 |
Translocations: 10
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr6 | 5112478 | Chr2 | 31527469 | |
| Translocation | Chr6 | 5112487 | Chr2 | 31584956 | |
| Translocation | Chr8 | 14194875 | Chr2 | 31584961 | LOC_Os08g23440 |
| Translocation | Chr8 | 14194876 | Chr2 | 31585415 | LOC_Os08g23440 |
| Translocation | Chr9 | 16386839 | Chr6 | 1605398 | LOC_Os06g28770 |
| Translocation | Chr10 | 18940554 | Chr8 | 13905772 | LOC_Os10g35436 |
| Translocation | Chr10 | 19785985 | Chr5 | 29565021 | |
| Translocation | Chr10 | 19802011 | Chr5 | 29565222 | LOC_Os10g36950 |
| Translocation | Chr4 | 22459334 | Chr2 | 31508234 | LOC_Os02g37150 |
| Translocation | Chr4 | 22459454 | Chr2 | 31527467 | LOC_Os02g37150 |
