Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN4490-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN4490-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 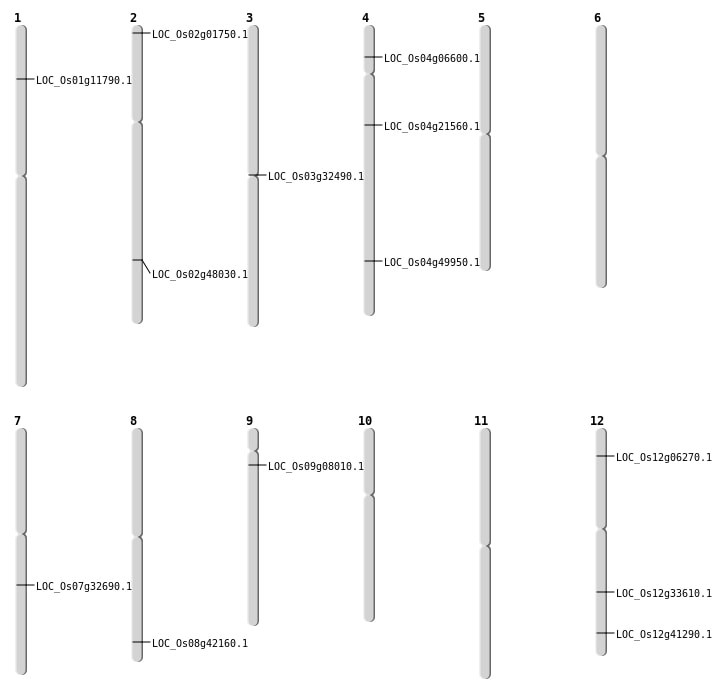
|
| Variant Information |
|---|
Single base substitutions: 33
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 11357561 | G-A | HOMO | INTERGENIC | |
| SBS | Chr1 | 6377301 | T-G | HET | NON_SYNONYMOUS_CODING | LOC_Os01g11790 |
| SBS | Chr10 | 21759404 | C-T | HET | INTRON | |
| SBS | Chr10 | 4481505 | C-T | HET | INTERGENIC | |
| SBS | Chr10 | 8469693 | A-G | HOMO | INTERGENIC | |
| SBS | Chr11 | 10556348 | G-T | HET | INTERGENIC | |
| SBS | Chr12 | 15379598 | C-A | HOMO | INTERGENIC | |
| SBS | Chr12 | 20229735 | C-T | HET | INTERGENIC | |
| SBS | Chr12 | 20229736 | C-T | HET | INTERGENIC | |
| SBS | Chr12 | 20293277 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os12g33610 |
| SBS | Chr12 | 24962161 | C-T | HET | SYNONYMOUS_CODING | |
| SBS | Chr12 | 26416650 | C-T | HET | SYNONYMOUS_CODING | |
| SBS | Chr2 | 18978404 | G-A | HOMO | INTERGENIC | |
| SBS | Chr2 | 2456486 | G-A | HOMO | INTERGENIC | |
| SBS | Chr2 | 29401125 | G-C | HOMO | NON_SYNONYMOUS_CODING | LOC_Os02g48030 |
| SBS | Chr3 | 1241261 | T-A | HET | INTRON | |
| SBS | Chr3 | 6543356 | T-A | HOMO | INTERGENIC | |
| SBS | Chr4 | 15990182 | C-A | HOMO | INTERGENIC | |
| SBS | Chr4 | 26205175 | T-A | HET | INTERGENIC | |
| SBS | Chr4 | 29618440 | T-A | HOMO | INTERGENIC | |
| SBS | Chr4 | 29618441 | A-T | HOMO | INTERGENIC | |
| SBS | Chr4 | 29793493 | C-T | HOMO | NON_SYNONYMOUS_CODING | LOC_Os04g49950 |
| SBS | Chr4 | 5413471 | G-A | HET | INTERGENIC | |
| SBS | Chr6 | 26995962 | C-T | HET | INTERGENIC | |
| SBS | Chr7 | 19522071 | A-C | HET | NON_SYNONYMOUS_CODING | LOC_Os07g32690 |
| SBS | Chr8 | 12627368 | G-T | HET | INTRON | |
| SBS | Chr8 | 15232390 | T-C | HET | INTERGENIC | |
| SBS | Chr8 | 18522723 | C-T | HET | INTERGENIC | |
| SBS | Chr8 | 19295023 | T-A | HET | INTERGENIC | |
| SBS | Chr8 | 26645490 | C-A | HET | NON_SYNONYMOUS_CODING | LOC_Os08g42160 |
| SBS | Chr8 | 5814561 | T-A | HET | INTRON | |
| SBS | Chr8 | 5845981 | T-A | HET | INTERGENIC | |
| SBS | Chr9 | 16457551 | T-A | HET | INTRON |
Deletions: 39
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr2 | 419940 | 419958 | 18 | LOC_Os02g01750 |
| Deletion | Chr10 | 568481 | 568534 | 53 | |
| Deletion | Chr12 | 1097898 | 1097909 | 11 | |
| Deletion | Chr8 | 2539084 | 2539088 | 4 | |
| Deletion | Chr12 | 2990177 | 2990179 | 2 | LOC_Os12g06270 |
| Deletion | Chr4 | 3486929 | 3486935 | 6 | |
| Deletion | Chr3 | 3610836 | 3610837 | 1 | |
| Deletion | Chr8 | 4273561 | 4273565 | 4 | |
| Deletion | Chr9 | 5765821 | 5765822 | 1 | |
| Deletion | Chr4 | 9643602 | 9643604 | 2 | |
| Deletion | Chr9 | 10205057 | 10205068 | 11 | |
| Deletion | Chr1 | 11444117 | 11444123 | 6 | |
| Deletion | Chr7 | 11963133 | 11963139 | 6 | |
| Deletion | Chr10 | 11989368 | 11989372 | 4 | |
| Deletion | Chr5 | 12084591 | 12084598 | 7 | |
| Deletion | Chr4 | 12187785 | 12187786 | 1 | LOC_Os04g21560 |
| Deletion | Chr8 | 13732161 | 13732169 | 8 | |
| Deletion | Chr10 | 16241276 | 16241279 | 3 | |
| Deletion | Chr4 | 17102396 | 17102397 | 1 | |
| Deletion | Chr4 | 18147648 | 18147654 | 6 | |
| Deletion | Chr12 | 19652252 | 19652261 | 9 | |
| Deletion | Chr12 | 21448731 | 21448732 | 1 | |
| Deletion | Chr6 | 21931023 | 21931024 | 1 | |
| Deletion | Chr4 | 22688539 | 22688540 | 1 | |
| Deletion | Chr1 | 23075416 | 23075419 | 3 | |
| Deletion | Chr6 | 23438249 | 23438250 | 1 | |
| Deletion | Chr8 | 23472998 | 23472999 | 1 | |
| Deletion | Chr12 | 23802903 | 23802923 | 20 | |
| Deletion | Chr12 | 25608275 | 25608277 | 2 | LOC_Os12g41290 |
| Deletion | Chr5 | 27070333 | 27070336 | 3 | |
| Deletion | Chr6 | 27466835 | 27466836 | 1 | |
| Deletion | Chr1 | 31135548 | 31135576 | 28 | |
| Deletion | Chr1 | 31508313 | 31508314 | 1 | |
| Deletion | Chr2 | 32415862 | 32415863 | 1 | |
| Deletion | Chr1 | 33598893 | 33598901 | 8 | |
| Deletion | Chr2 | 34581468 | 34581469 | 1 | |
| Deletion | Chr4 | 34728219 | 34728220 | 1 | |
| Deletion | Chr1 | 37285892 | 37285893 | 1 | |
| Deletion | Chr1 | 38039042 | 38039060 | 18 |
Insertions: 12
Inversions: 2
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr12 | 5687843 | 5687981 | |
| Inversion | Chr11 | 13549026 | 13549229 |
Translocations: 5
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr9 | 3480180 | Chr4 | 11078317 | LOC_Os04g06600 |
| Translocation | Chr12 | 4126479 | Chr9 | 5123547 | LOC_Os09g08010 |
| Translocation | Chr4 | 18577003 | Chr3 | 34252811 | LOC_Os03g32490 |
| Translocation | Chr3 | 34696783 | Chr2 | 33958058 | |
| Translocation | Chr3 | 34696998 | Chr2 | 33958051 |
