Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN4491-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN4491-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 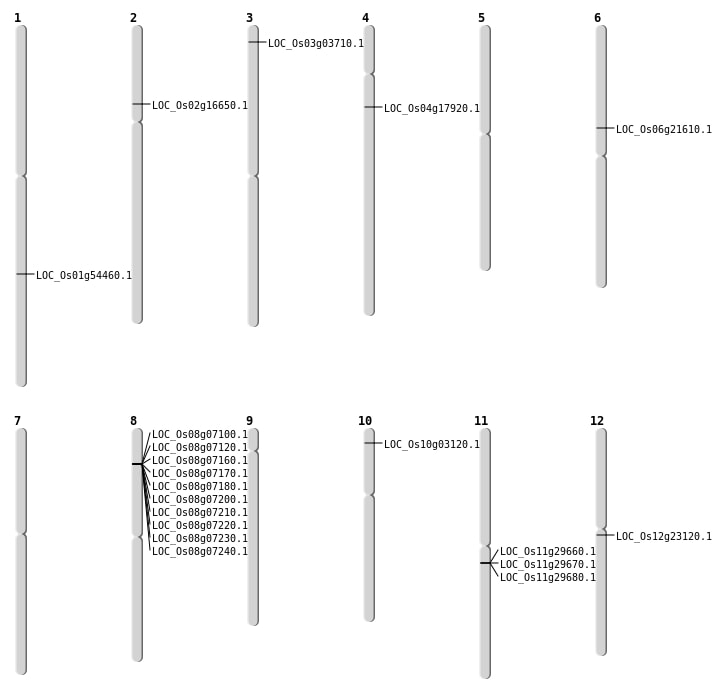
|
| Variant Information |
|---|
Single base substitutions: 40
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 17535829 | C-T | HOMO | INTRON | |
| SBS | Chr1 | 22511530 | G-A | HET | INTERGENIC | |
| SBS | Chr1 | 24144971 | A-G | HET | INTERGENIC | |
| SBS | Chr1 | 31328083 | A-T | HET | NON_SYNONYMOUS_CODING | LOC_Os01g54460 |
| SBS | Chr1 | 33956433 | C-G | HET | INTERGENIC | |
| SBS | Chr1 | 42107512 | C-T | HET | INTERGENIC | |
| SBS | Chr10 | 15566200 | C-T | HET | SYNONYMOUS_CODING | |
| SBS | Chr11 | 21852097 | T-C | HET | INTERGENIC | |
| SBS | Chr12 | 13074504 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os12g23120 |
| SBS | Chr12 | 21792834 | G-C | HET | INTERGENIC | |
| SBS | Chr12 | 25879052 | A-C | HOMO | INTERGENIC | |
| SBS | Chr2 | 10639058 | C-A | HOMO | INTRON | |
| SBS | Chr2 | 30470771 | C-T | HET | INTERGENIC | |
| SBS | Chr2 | 30817327 | A-T | HET | INTERGENIC | |
| SBS | Chr2 | 32636911 | A-T | HET | INTERGENIC | |
| SBS | Chr2 | 9511257 | C-A | HET | NON_SYNONYMOUS_CODING | LOC_Os02g16650 |
| SBS | Chr3 | 13787145 | G-A | HET | UTR_3_PRIME | |
| SBS | Chr3 | 16142843 | C-T | HOMO | INTERGENIC | |
| SBS | Chr3 | 1634416 | T-C | HOMO | INTRON | |
| SBS | Chr3 | 1653532 | C-G | HOMO | NON_SYNONYMOUS_CODING | LOC_Os03g03710 |
| SBS | Chr3 | 18962227 | G-A | HOMO | INTERGENIC | |
| SBS | Chr3 | 19813059 | A-G | HET | INTERGENIC | |
| SBS | Chr3 | 19961631 | C-A | HET | INTERGENIC | |
| SBS | Chr3 | 35293321 | C-T | HET | INTRON | |
| SBS | Chr3 | 9455034 | C-G | HOMO | UTR_3_PRIME | |
| SBS | Chr3 | 956380 | C-T | HET | SYNONYMOUS_CODING | |
| SBS | Chr5 | 10197746 | G-A | HET | INTERGENIC | |
| SBS | Chr5 | 16075576 | G-A | HOMO | SYNONYMOUS_CODING | |
| SBS | Chr5 | 25080659 | G-T | HOMO | INTERGENIC | |
| SBS | Chr5 | 3038282 | T-C | HET | INTERGENIC | |
| SBS | Chr5 | 3817892 | T-A | HET | INTRON | |
| SBS | Chr6 | 10867562 | C-T | HET | SYNONYMOUS_CODING | |
| SBS | Chr6 | 12469871 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os06g21610 |
| SBS | Chr6 | 16037593 | G-A | HET | INTERGENIC | |
| SBS | Chr6 | 16332207 | G-A | HET | INTERGENIC | |
| SBS | Chr7 | 10967038 | G-A | HOMO | INTRON | |
| SBS | Chr7 | 21111650 | C-T | HOMO | INTERGENIC | |
| SBS | Chr8 | 16486642 | T-C | HET | INTERGENIC | |
| SBS | Chr8 | 9318713 | G-A | HET | INTERGENIC | |
| SBS | Chr9 | 11379268 | C-T | HET | INTERGENIC |
Deletions: 30
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr7 | 531104 | 531105 | 1 | |
| Deletion | Chr10 | 1572764 | 1572765 | 1 | |
| Deletion | Chr1 | 1749614 | 1749616 | 2 | |
| Deletion | Chr3 | 3059092 | 3059101 | 9 | |
| Deletion | Chr7 | 3462309 | 3462323 | 14 | |
| Deletion | Chr8 | 3976001 | 3991000 | 15000 | 2 |
| Deletion | Chr8 | 4003001 | 4024000 | 21000 | 6 |
| Deletion | Chr8 | 4028001 | 4050000 | 22000 | 3 |
| Deletion | Chr5 | 4081461 | 4081462 | 1 | |
| Deletion | Chr8 | 4163159 | 4163161 | 2 | |
| Deletion | Chr7 | 4242972 | 4242974 | 2 | |
| Deletion | Chr4 | 9854056 | 9854133 | 77 | LOC_Os04g17920 |
| Deletion | Chr2 | 13894634 | 13894636 | 2 | |
| Deletion | Chr2 | 14032541 | 14032563 | 22 | |
| Deletion | Chr1 | 15415060 | 15415061 | 1 | |
| Deletion | Chr4 | 15537147 | 15537148 | 1 | |
| Deletion | Chr2 | 16341287 | 16341289 | 2 | |
| Deletion | Chr4 | 16605497 | 16605498 | 1 | |
| Deletion | Chr7 | 17111742 | 17111743 | 1 | |
| Deletion | Chr11 | 17193001 | 17208000 | 15000 | 3 |
| Deletion | Chr10 | 17392605 | 17392628 | 23 | |
| Deletion | Chr8 | 19997767 | 19997769 | 2 | |
| Deletion | Chr4 | 20059535 | 20059536 | 1 | |
| Deletion | Chr3 | 21574734 | 21574735 | 1 | |
| Deletion | Chr1 | 21933089 | 21933094 | 5 | |
| Deletion | Chr1 | 22593585 | 22593586 | 1 | |
| Deletion | Chr12 | 25060020 | 25060021 | 1 | |
| Deletion | Chr4 | 27274503 | 27274505 | 2 | |
| Deletion | Chr2 | 32437292 | 32437296 | 4 | |
| Deletion | Chr2 | 34694784 | 34694790 | 6 |
Insertions: 8
Inversions: 3
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr10 | 1270806 | 1318610 | LOC_Os10g03120 |
| Inversion | Chr10 | 2276142 | 2276247 | |
| Inversion | Chr8 | 21563824 | 21563950 |
Translocations: 2
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr8 | 21275227 | Chr1 | 39382861 | |
| Translocation | Chr12 | 25890388 | Chr6 | 353816 |
