Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN4492-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN4492-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 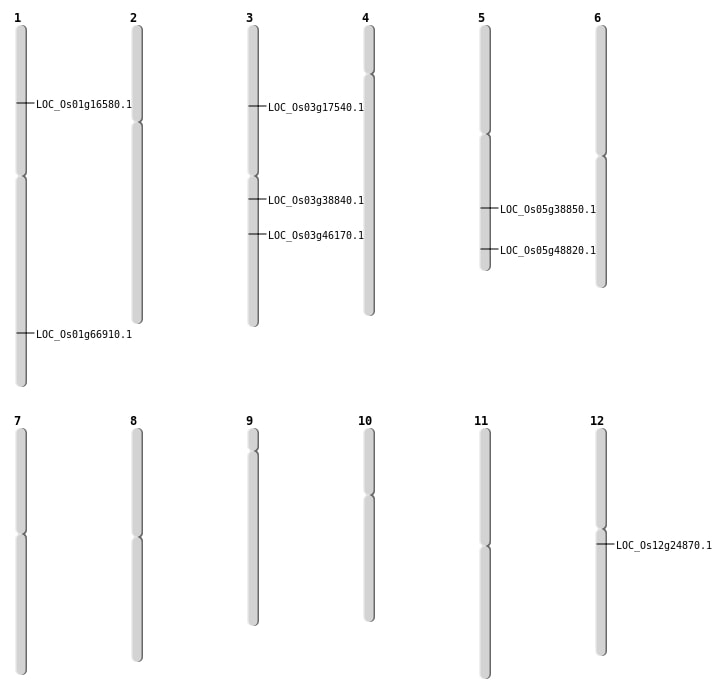
|
| Variant Information |
|---|
Single base substitutions: 36
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 18192317 | C-A | HOMO | INTRON | |
| SBS | Chr1 | 18845021 | C-T | HOMO | INTERGENIC | |
| SBS | Chr1 | 38860088 | G-T | HET | NON_SYNONYMOUS_CODING | LOC_Os01g66910 |
| SBS | Chr1 | 8224765 | T-C | HET | INTERGENIC | |
| SBS | Chr1 | 9414716 | A-T | HET | NON_SYNONYMOUS_CODING | LOC_Os01g16580 |
| SBS | Chr10 | 257609 | C-T | HET | INTERGENIC | |
| SBS | Chr11 | 12727519 | G-T | HET | INTRON | |
| SBS | Chr11 | 13131038 | T-G | HET | INTERGENIC | |
| SBS | Chr11 | 8152456 | G-A | HET | INTRON | |
| SBS | Chr12 | 14259175 | C-A | HOMO | NON_SYNONYMOUS_CODING | LOC_Os12g24870 |
| SBS | Chr12 | 1616012 | A-T | HET | INTRON | |
| SBS | Chr12 | 8980445 | C-T | HOMO | INTERGENIC | |
| SBS | Chr2 | 336414 | A-G | HOMO | INTERGENIC | |
| SBS | Chr2 | 837741 | T-A | HOMO | INTERGENIC | |
| SBS | Chr3 | 21570120 | A-G | HET | NON_SYNONYMOUS_CODING | LOC_Os03g38840 |
| SBS | Chr3 | 21830678 | C-T | HET | INTRON | |
| SBS | Chr3 | 25837312 | T-C | HET | INTERGENIC | |
| SBS | Chr3 | 26103093 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os03g46170 |
| SBS | Chr3 | 29154654 | C-T | HET | INTERGENIC | |
| SBS | Chr3 | 2922510 | C-T | HET | INTRON | |
| SBS | Chr3 | 31571915 | G-A | HET | INTERGENIC | |
| SBS | Chr4 | 29815324 | G-A | HET | INTRON | |
| SBS | Chr5 | 15218839 | C-T | HET | INTRON | |
| SBS | Chr5 | 15218843 | G-T | HET | INTRON | |
| SBS | Chr5 | 27979829 | T-G | HET | NON_SYNONYMOUS_CODING | LOC_Os05g48820 |
| SBS | Chr5 | 29903555 | G-A | HET | INTRON | |
| SBS | Chr5 | 5170589 | A-T | HET | INTERGENIC | |
| SBS | Chr6 | 20188829 | T-C | HET | INTERGENIC | |
| SBS | Chr6 | 20936045 | A-G | HET | INTERGENIC | |
| SBS | Chr6 | 2383059 | G-A | HET | INTERGENIC | |
| SBS | Chr7 | 18385200 | T-C | HOMO | SYNONYMOUS_CODING | |
| SBS | Chr7 | 184007 | G-A | HET | INTERGENIC | |
| SBS | Chr7 | 20689103 | A-G | HET | SPLICE_SITE_REGION | |
| SBS | Chr8 | 19039592 | A-T | HOMO | INTERGENIC | |
| SBS | Chr9 | 12398363 | T-C | HOMO | UTR_5_PRIME | |
| SBS | Chr9 | 2307505 | A-G | HOMO | INTERGENIC |
Deletions: 25
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr2 | 111180 | 111181 | 1 | |
| Deletion | Chr7 | 370539 | 370549 | 10 | |
| Deletion | Chr11 | 738113 | 738122 | 9 | |
| Deletion | Chr5 | 1431760 | 1431761 | 1 | |
| Deletion | Chr9 | 2055218 | 2055219 | 1 | |
| Deletion | Chr3 | 6037212 | 6037218 | 6 | |
| Deletion | Chr2 | 7367001 | 7380000 | 13000 | |
| Deletion | Chr7 | 7601329 | 7601330 | 1 | |
| Deletion | Chr11 | 7947681 | 7947688 | 7 | |
| Deletion | Chr7 | 9727317 | 9727319 | 2 | |
| Deletion | Chr9 | 11273397 | 11273403 | 6 | |
| Deletion | Chr4 | 11889546 | 11889547 | 1 | |
| Deletion | Chr9 | 14005365 | 14005379 | 14 | |
| Deletion | Chr11 | 14542477 | 14542478 | 1 | |
| Deletion | Chr3 | 20230098 | 20230099 | 1 | |
| Deletion | Chr4 | 21221994 | 21221995 | 1 | |
| Deletion | Chr3 | 22540594 | 22540595 | 1 | |
| Deletion | Chr5 | 22790489 | 22790490 | 1 | LOC_Os05g38850 |
| Deletion | Chr12 | 23090899 | 23090919 | 20 | |
| Deletion | Chr7 | 23871034 | 23871041 | 7 | |
| Deletion | Chr11 | 27170986 | 27170987 | 1 | |
| Deletion | Chr8 | 27871057 | 27871058 | 1 | |
| Deletion | Chr1 | 30349325 | 30349326 | 1 | |
| Deletion | Chr4 | 32893831 | 32893832 | 1 | |
| Deletion | Chr4 | 34462528 | 34462529 | 1 |
Insertions: 11
Inversions: 3
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr8 | 8015595 | 8015809 | |
| Inversion | Chr9 | 18014348 | 18014530 | |
| Inversion | Chr3 | 27564363 | 27564475 |
Translocations: 2
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr8 | 9744461 | Chr3 | 16641765 | LOC_Os03g17540 |
| Translocation | Chr6 | 28880844 | Chr2 | 7385049 |
