Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN4493-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN4493-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 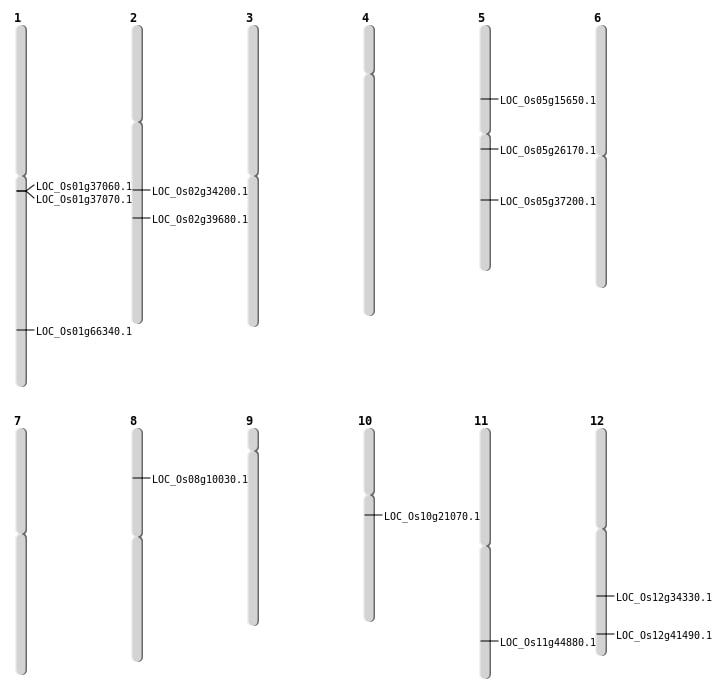
|
| Variant Information |
|---|
Single base substitutions: 44
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 13970714 | T-C | HET | INTERGENIC | |
| SBS | Chr1 | 19029783 | T-G | HET | INTERGENIC | |
| SBS | Chr1 | 1976098 | C-T | HOMO | INTRON | |
| SBS | Chr1 | 38528048 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os01g66340 |
| SBS | Chr10 | 10668946 | G-C | HET | NON_SYNONYMOUS_CODING | LOC_Os10g21070 |
| SBS | Chr10 | 12447143 | C-T | HET | INTERGENIC | |
| SBS | Chr10 | 1319694 | A-C | HET | INTRON | |
| SBS | Chr10 | 6868072 | C-A | HET | INTERGENIC | |
| SBS | Chr10 | 9278142 | T-A | HET | INTERGENIC | |
| SBS | Chr10 | 9405944 | T-G | HET | INTERGENIC | |
| SBS | Chr11 | 18882866 | C-A | HET | INTERGENIC | |
| SBS | Chr11 | 22011814 | T-C | HET | INTERGENIC | |
| SBS | Chr11 | 27163444 | A-G | HOMO | NON_SYNONYMOUS_CODING | LOC_Os11g44880 |
| SBS | Chr12 | 19295343 | A-T | HET | INTERGENIC | |
| SBS | Chr12 | 25701245 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os12g41490 |
| SBS | Chr12 | 4162275 | A-T | HET | INTERGENIC | |
| SBS | Chr2 | 18738977 | A-G | HOMO | INTERGENIC | |
| SBS | Chr2 | 20222182 | G-T | HET | SYNONYMOUS_CODING | |
| SBS | Chr2 | 20460689 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os02g34200 |
| SBS | Chr2 | 20460690 | C-A | HET | SYNONYMOUS_CODING | |
| SBS | Chr2 | 24967731 | C-A | HET | INTERGENIC | |
| SBS | Chr2 | 28466677 | C-T | HET | INTRON | |
| SBS | Chr2 | 34022594 | A-G | HET | INTERGENIC | |
| SBS | Chr2 | 6544284 | G-A | HET | INTERGENIC | |
| SBS | Chr3 | 19451399 | C-G | HET | INTERGENIC | |
| SBS | Chr3 | 19451400 | G-T | HET | INTERGENIC | |
| SBS | Chr3 | 33677307 | G-T | HET | INTERGENIC | |
| SBS | Chr3 | 5862478 | A-C | HOMO | INTRON | |
| SBS | Chr3 | 8719072 | A-G | HET | SYNONYMOUS_CODING | |
| SBS | Chr4 | 13676260 | G-T | HET | INTERGENIC | |
| SBS | Chr4 | 14145340 | C-T | HOMO | INTERGENIC | |
| SBS | Chr4 | 16660052 | C-G | HET | INTERGENIC | |
| SBS | Chr4 | 56602 | T-C | HET | INTERGENIC | |
| SBS | Chr5 | 12642332 | C-T | HET | SYNONYMOUS_CODING | |
| SBS | Chr5 | 15230673 | C-G | HET | NON_SYNONYMOUS_CODING | LOC_Os05g26170 |
| SBS | Chr6 | 22114513 | G-A | HET | INTERGENIC | |
| SBS | Chr6 | 9624022 | G-C | HET | INTERGENIC | |
| SBS | Chr7 | 11752589 | T-C | HET | INTERGENIC | |
| SBS | Chr7 | 14274153 | G-A | HET | INTERGENIC | |
| SBS | Chr7 | 6200828 | A-G | HET | INTERGENIC | |
| SBS | Chr9 | 12160029 | T-C | HET | INTRON | |
| SBS | Chr9 | 14528450 | G-A | HET | INTERGENIC | |
| SBS | Chr9 | 1568902 | A-G | HET | SYNONYMOUS_CODING | |
| SBS | Chr9 | 7059075 | T-C | HET | INTERGENIC |
Deletions: 19
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr6 | 1212397 | 1212398 | 1 | |
| Deletion | Chr4 | 2872949 | 2873059 | 110 | |
| Deletion | Chr5 | 6613721 | 6613722 | 1 | |
| Deletion | Chr4 | 8576407 | 8576425 | 18 | |
| Deletion | Chr10 | 9683082 | 9683129 | 47 | |
| Deletion | Chr11 | 11380109 | 11380110 | 1 | |
| Deletion | Chr7 | 12589643 | 12589645 | 2 | |
| Deletion | Chr1 | 14981503 | 14981504 | 1 | |
| Deletion | Chr7 | 15404951 | 15404952 | 1 | |
| Deletion | Chr6 | 19641970 | 19641971 | 1 | |
| Deletion | Chr1 | 20674001 | 20690000 | 16000 | 2 |
| Deletion | Chr12 | 20807846 | 20807855 | 9 | LOC_Os12g34330 |
| Deletion | Chr9 | 20913975 | 20913988 | 13 | |
| Deletion | Chr10 | 21672549 | 21672550 | 1 | |
| Deletion | Chr9 | 21686911 | 21686918 | 7 | |
| Deletion | Chr2 | 23971362 | 23971439 | 77 | LOC_Os02g39680 |
| Deletion | Chr12 | 24649406 | 24649419 | 13 | |
| Deletion | Chr6 | 29334764 | 29334774 | 10 | |
| Deletion | Chr3 | 29621139 | 29621141 | 2 |
Insertions: 21
Inversions: 5
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr8 | 5805035 | 5814014 | LOC_Os08g10030 |
| Inversion | Chr8 | 5805048 | 5814015 | LOC_Os08g10030 |
| Inversion | Chr3 | 11374266 | 11558763 | |
| Inversion | Chr7 | 13021517 | 13021568 | |
| Inversion | Chr5 | 21753476 | 21753906 | LOC_Os05g37200 |
Translocations: 6
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr5 | 8852674 | Chr1 | 37211317 | LOC_Os05g15650 |
| Translocation | Chr3 | 11611320 | Chr1 | 42094216 | |
| Translocation | Chr6 | 12360285 | Chr3 | 11611056 | |
| Translocation | Chr6 | 12360502 | Chr1 | 40966577 | |
| Translocation | Chr7 | 12785163 | Chr6 | 12360301 | |
| Translocation | Chr7 | 12785164 | Chr6 | 12360502 |
