Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN450-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN450-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 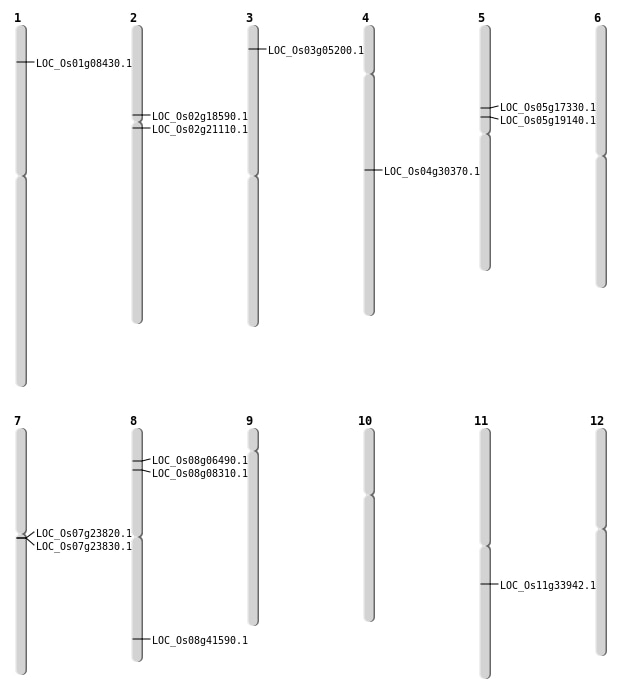
|
| Variant Information |
|---|
Single base substitutions: 26
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 14567016 | G-T | HET | INTERGENIC | |
| SBS | Chr1 | 18368594 | T-C | HOMO | INTERGENIC | |
| SBS | Chr1 | 25821813 | T-G | HOMO | INTERGENIC | |
| SBS | Chr1 | 27136599 | A-G | HOMO | INTERGENIC | |
| SBS | Chr1 | 4143624 | T-C | HOMO | NON_SYNONYMOUS_CODING | LOC_Os01g08430 |
| SBS | Chr10 | 10117495 | C-T | HOMO | INTERGENIC | |
| SBS | Chr10 | 19558414 | T-C | HET | UTR_3_PRIME | |
| SBS | Chr11 | 4510373 | A-C | HOMO | INTERGENIC | |
| SBS | Chr11 | 8578616 | G-A | HOMO | INTERGENIC | |
| SBS | Chr12 | 18007261 | A-T | HET | INTRON | |
| SBS | Chr12 | 6748174 | G-C | HET | INTERGENIC | |
| SBS | Chr3 | 15653906 | A-G | HOMO | INTERGENIC | |
| SBS | Chr3 | 2525957 | A-C | HET | START_GAINED | LOC_Os03g05200 |
| SBS | Chr3 | 2919083 | T-A | HET | INTRON | |
| SBS | Chr3 | 3000453 | A-T | HET | INTERGENIC | |
| SBS | Chr4 | 14463856 | A-G | HET | INTERGENIC | |
| SBS | Chr4 | 18071611 | C-G | HET | SYNONYMOUS_CODING | |
| SBS | Chr5 | 25073339 | G-A | HOMO | INTERGENIC | |
| SBS | Chr6 | 26882587 | C-T | HET | INTERGENIC | |
| SBS | Chr6 | 8174468 | C-G | HET | INTERGENIC | |
| SBS | Chr7 | 2826055 | A-G | HOMO | INTRON | |
| SBS | Chr8 | 13204633 | G-T | HET | INTRON | |
| SBS | Chr8 | 18238311 | C-T | HOMO | INTERGENIC | |
| SBS | Chr8 | 23179860 | G-A | HET | INTERGENIC | |
| SBS | Chr9 | 14439781 | G-C | HOMO | SYNONYMOUS_CODING | |
| SBS | Chr9 | 17797063 | C-T | HET | INTERGENIC |
Deletions: 11
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr3 | 731128 | 731135 | 7 | |
| Deletion | Chr8 | 4767001 | 4778000 | 11000 | LOC_Os08g08310 |
| Deletion | Chr9 | 5462238 | 5462249 | 11 | |
| Deletion | Chr10 | 9578246 | 9578254 | 8 | |
| Deletion | Chr2 | 10618359 | 10618365 | 6 | |
| Deletion | Chr2 | 10841919 | 10841920 | 1 | LOC_Os02g18590 |
| Deletion | Chr5 | 11108927 | 11108938 | 11 | LOC_Os05g19140 |
| Deletion | Chr7 | 13452001 | 13460000 | 8000 | 2 |
| Deletion | Chr11 | 19196961 | 19196976 | 15 | |
| Deletion | Chr3 | 29046619 | 29046622 | 3 | |
| Deletion | Chr3 | 30454514 | 30454518 | 4 |
No Insertion
Inversions: 2
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr8 | 4777675 | 26270040 | LOC_Os08g41590 |
| Inversion | Chr8 | 4778138 | 26270043 | LOC_Os08g41590 |
Translocations: 9
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr10 | 158659 | Chr2 | 19267959 | |
| Translocation | Chr8 | 3586919 | Chr4 | 7627822 | |
| Translocation | Chr8 | 3678739 | Chr4 | 7628170 | LOC_Os08g06490 |
| Translocation | Chr5 | 9934647 | Chr2 | 12529488 | 2 |
| Translocation | Chr4 | 18124188 | Chr1 | 9642782 | LOC_Os04g30370 |
| Translocation | Chr6 | 18200663 | Chr2 | 8950274 | |
| Translocation | Chr11 | 19839735 | Chr4 | 2842007 | LOC_Os11g33942 |
| Translocation | Chr11 | 21846778 | Chr6 | 25106499 | |
| Translocation | Chr11 | 21846792 | Chr6 | 25106223 |
