Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN4505-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN4505-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 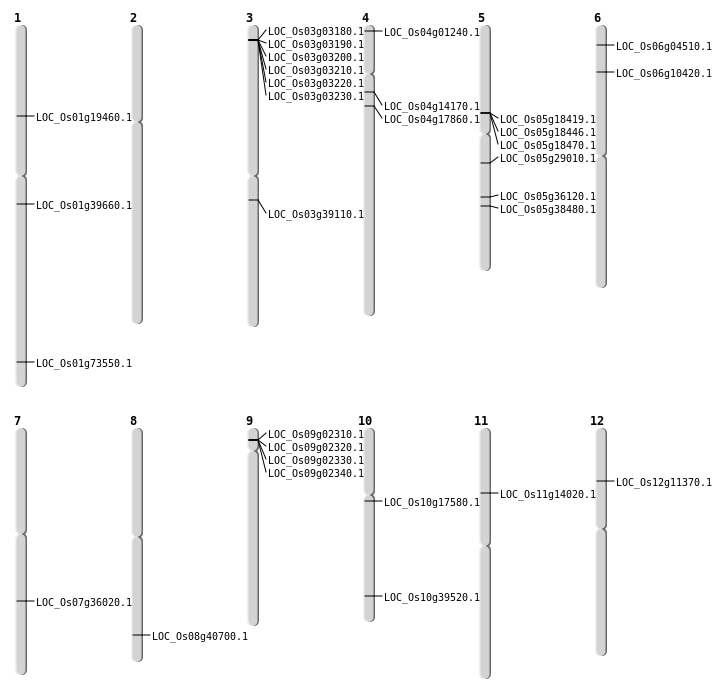
|
| Variant Information |
|---|
Single base substitutions: 42
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 26137219 | C-T | HOMO | ||
| SBS | Chr1 | 27386438 | A-T | HET | ||
| SBS | Chr1 | 30727661 | T-C | HET | ||
| SBS | Chr1 | 31805080 | C-T | HOMO | ||
| SBS | Chr1 | 35421302 | C-T | HET | ||
| SBS | Chr1 | 42046936 | C-T | HET | ||
| SBS | Chr1 | 6122593 | G-A | HOMO | ||
| SBS | Chr10 | 12824281 | G-A | HOMO | ||
| SBS | Chr10 | 1953673 | C-T | HOMO | ||
| SBS | Chr10 | 3680774 | A-G | HET | ||
| SBS | Chr10 | 8880885 | T-A | HET | LOC_Os10g17580 | |
| SBS | Chr11 | 10402664 | T-C | HET | ||
| SBS | Chr11 | 12289225 | C-G | HET | ||
| SBS | Chr12 | 11294603 | G-A | HET | ||
| SBS | Chr12 | 22512475 | A-G | HET | ||
| SBS | Chr2 | 22278482 | G-T | HET | ||
| SBS | Chr3 | 10021080 | A-C | HET | ||
| SBS | Chr3 | 16385681 | G-A | HET | ||
| SBS | Chr3 | 26423174 | C-T | HET | ||
| SBS | Chr3 | 689675 | T-A | HET | ||
| SBS | Chr4 | 7935106 | T-C | HET | LOC_Os04g14170 | |
| SBS | Chr5 | 1021507 | T-C | HET | ||
| SBS | Chr5 | 10428979 | C-A | HET | ||
| SBS | Chr5 | 13014073 | G-A | HET | ||
| SBS | Chr5 | 14227545 | T-A | HET | ||
| SBS | Chr5 | 16513637 | A-G | HET | ||
| SBS | Chr5 | 17009396 | C-A | HET | LOC_Os05g29010 | |
| SBS | Chr5 | 9465746 | A-G | HET | ||
| SBS | Chr6 | 13784064 | G-A | HET | ||
| SBS | Chr6 | 1936034 | C-T | HET | LOC_Os06g04510 | |
| SBS | Chr6 | 1980078 | C-T | HET | ||
| SBS | Chr6 | 25245521 | T-C | HET | ||
| SBS | Chr6 | 26044842 | G-A | HET | ||
| SBS | Chr7 | 24769519 | C-A | HET | ||
| SBS | Chr8 | 11741898 | T-G | HET | ||
| SBS | Chr8 | 12899670 | C-T | HET | ||
| SBS | Chr8 | 19987271 | T-A | HET | ||
| SBS | Chr8 | 25766038 | G-T | HET | LOC_Os08g40700 | |
| SBS | Chr8 | 6568996 | A-T | HET | ||
| SBS | Chr9 | 19706907 | A-G | HET | ||
| SBS | Chr9 | 4266807 | G-A | HET | ||
| SBS | Chr9 | 8980663 | C-T | HET |
Deletions: 45
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr3 | 42353 | 42353 | 1 | |
| Deletion | Chr4 | 182962 | 182963 | 2 | LOC_Os04g01240 |
| Deletion | Chr8 | 407645 | 407645 | 1 | |
| Deletion | Chr5 | 778443 | 778449 | 7 | |
| Deletion | Chr9 | 940001 | 967000 | 27000 | 4 |
| Deletion | Chr3 | 1347001 | 1381000 | 34000 | 6 |
| Deletion | Chr3 | 1364795 | 1364795 | 1 | LOC_Os03g03210 |
| Deletion | Chr4 | 2063256 | 2063256 | 1 | |
| Deletion | Chr1 | 2748084 | 2748085 | 2 | |
| Deletion | Chr4 | 2891575 | 2891580 | 6 | |
| Deletion | Chr9 | 4802996 | 4802997 | 2 | |
| Deletion | Chr2 | 4994700 | 4994700 | 1 | |
| Deletion | Chr6 | 5357116 | 5357116 | 1 | LOC_Os06g10420 |
| Deletion | Chr5 | 5566063 | 5566140 | 78 | |
| Deletion | Chr12 | 6527443 | 6527523 | 81 | |
| Deletion | Chr3 | 7489098 | 7489098 | 1 | |
| Deletion | Chr4 | 9003268 | 9003268 | 1 | |
| Deletion | Chr4 | 9810435 | 9810435 | 1 | LOC_Os04g17860 |
| Deletion | Chr8 | 10547302 | 10547302 | 1 | |
| Deletion | Chr5 | 10652001 | 10676000 | 24000 | 3 |
| Deletion | Chr12 | 10924831 | 10924831 | 1 | |
| Deletion | Chr1 | 11016491 | 11016512 | 22 | LOC_Os01g19460 |
| Deletion | Chr3 | 11232292 | 11232292 | 1 | |
| Deletion | Chr6 | 11416570 | 11416570 | 1 | |
| Deletion | Chr12 | 11526896 | 11526896 | 1 | |
| Deletion | Chr5 | 11598476 | 11598478 | 3 | |
| Deletion | Chr3 | 13414166 | 13414240 | 75 | |
| Deletion | Chr9 | 13751090 | 13751090 | 1 | |
| Deletion | Chr5 | 16089860 | 16089860 | 1 | |
| Deletion | Chr1 | 16944540 | 16945179 | 640 | |
| Deletion | Chr11 | 17706471 | 17706498 | 28 | |
| Deletion | Chr3 | 18947478 | 18947480 | 3 | |
| Deletion | Chr12 | 18996140 | 18996140 | 1 | |
| Deletion | Chr1 | 19523459 | 19523459 | 1 | |
| Deletion | Chr1 | 20844058 | 20844063 | 6 | |
| Deletion | Chr7 | 21475653 | 21475653 | 1 | |
| Deletion | Chr5 | 22576568 | 22576568 | 1 | LOC_Os05g38480 |
| Deletion | Chr7 | 22849171 | 22849175 | 5 | |
| Deletion | Chr7 | 23874874 | 23874876 | 3 | |
| Deletion | Chr2 | 24240694 | 24240753 | 60 | |
| Deletion | Chr3 | 25433642 | 25433642 | 1 | |
| Deletion | Chr3 | 28086667 | 28086676 | 10 | |
| Deletion | Chr4 | 28317786 | 28317838 | 53 | |
| Deletion | Chr1 | 38874770 | 38874777 | 8 | |
| Deletion | Chr1 | 40899199 | 40899199 | 1 |
Insertions: 20
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr1 | 6944492 | 6944493 | 2 | |
| Insertion | Chr10 | 15983382 | 15983459 | 78 | |
| Insertion | Chr11 | 18224332 | 18224341 | 10 | |
| Insertion | Chr12 | 14093901 | 14093928 | 28 | |
| Insertion | Chr12 | 23505873 | 23505930 | 58 | |
| Insertion | Chr12 | 6139524 | 6139583 | 60 | LOC_Os12g11370 |
| Insertion | Chr12 | 8494395 | 8494395 | 1 | |
| Insertion | Chr2 | 14294048 | 14294050 | 3 | |
| Insertion | Chr2 | 23808168 | 23808168 | 1 | |
| Insertion | Chr3 | 33959989 | 33959990 | 2 | |
| Insertion | Chr4 | 29710501 | 29710501 | 1 | |
| Insertion | Chr4 | 31371702 | 31371790 | 89 | |
| Insertion | Chr6 | 19363020 | 19363109 | 90 | |
| Insertion | Chr6 | 4278038 | 4278162 | 125 | |
| Insertion | Chr8 | 13065401 | 13065401 | 1 | |
| Insertion | Chr8 | 14484024 | 14484071 | 48 | |
| Insertion | Chr8 | 3876340 | 3876361 | 22 | |
| Insertion | Chr8 | 5022341 | 5022343 | 3 | |
| Insertion | Chr8 | 5045957 | 5045957 | 1 | |
| Insertion | Chr8 | 5592424 | 5592429 | 6 |
Inversions: 2
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr1 | 42274519 | 42615373 | LOC_Os01g73550 |
| Inversion | Chr1 | 42274522 | 42615380 | LOC_Os01g73550 |
No Translocation
