Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN451-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN451-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 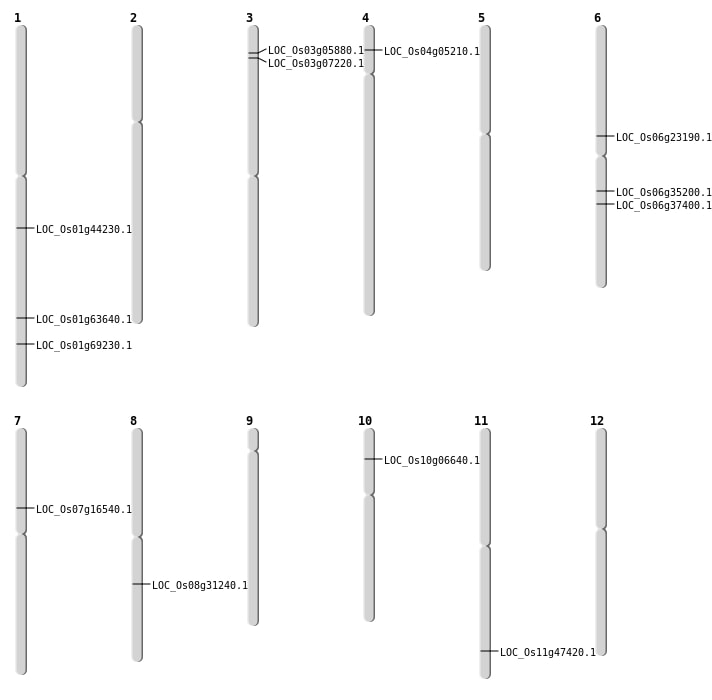
|
| Variant Information |
|---|
Single base substitutions: 36
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 11151066 | T-C | HOMO | SYNONYMOUS_CODING | |
| SBS | Chr1 | 22367559 | T-A | HOMO | SYNONYMOUS_CODING | |
| SBS | Chr1 | 35928555 | G-A | HOMO | INTRON | |
| SBS | Chr1 | 36704508 | C-T | HET | INTRON | |
| SBS | Chr1 | 36910778 | C-T | HOMO | NON_SYNONYMOUS_CODING | LOC_Os01g63640 |
| SBS | Chr1 | 39401050 | G-T | HOMO | INTERGENIC | |
| SBS | Chr11 | 3685285 | C-T | HET | INTERGENIC | |
| SBS | Chr11 | 6083727 | G-A | HOMO | INTERGENIC | |
| SBS | Chr12 | 1375704 | A-C | HOMO | INTERGENIC | |
| SBS | Chr2 | 10349716 | G-T | HET | INTERGENIC | |
| SBS | Chr2 | 10349717 | G-A | HET | INTERGENIC | |
| SBS | Chr3 | 14215063 | G-T | HET | INTRON | |
| SBS | Chr3 | 14215064 | A-T | HET | INTRON | |
| SBS | Chr3 | 15737573 | C-T | HET | INTERGENIC | |
| SBS | Chr3 | 20791654 | G-A | HET | INTERGENIC | |
| SBS | Chr3 | 7931377 | C-A | HOMO | INTRON | |
| SBS | Chr4 | 12273974 | T-A | HET | SYNONYMOUS_CODING | |
| SBS | Chr4 | 13103088 | C-T | HET | INTERGENIC | |
| SBS | Chr4 | 2602025 | T-G | HET | NON_SYNONYMOUS_CODING | LOC_Os04g05210 |
| SBS | Chr4 | 26340895 | C-T | HOMO | INTERGENIC | |
| SBS | Chr4 | 35214568 | G-T | HOMO | INTERGENIC | |
| SBS | Chr5 | 1030175 | A-G | HET | INTERGENIC | |
| SBS | Chr5 | 21701026 | A-G | HOMO | UTR_3_PRIME | |
| SBS | Chr5 | 29551958 | G-A | HET | INTERGENIC | |
| SBS | Chr5 | 6001172 | T-A | HET | INTERGENIC | |
| SBS | Chr6 | 10591957 | C-T | HET | INTERGENIC | |
| SBS | Chr6 | 13542075 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os06g23190 |
| SBS | Chr6 | 13542076 | T-G | HET | NON_SYNONYMOUS_CODING | LOC_Os06g23190 |
| SBS | Chr6 | 20524378 | C-A | HET | STOP_GAINED | LOC_Os06g35200 |
| SBS | Chr6 | 8920554 | A-T | HET | INTERGENIC | |
| SBS | Chr7 | 9016243 | C-T | HET | INTERGENIC | |
| SBS | Chr8 | 13808603 | C-T | HOMO | INTRON | |
| SBS | Chr8 | 18291393 | C-T | HOMO | INTERGENIC | |
| SBS | Chr8 | 18452807 | C-A | HOMO | UTR_5_PRIME | |
| SBS | Chr8 | 20116999 | C-T | HET | INTERGENIC | |
| SBS | Chr9 | 8186761 | C-T | HET | INTERGENIC |
Deletions: 24
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr12 | 961836 | 961849 | 13 | |
| Deletion | Chr11 | 1663830 | 1663834 | 4 | |
| Deletion | Chr11 | 2085565 | 2085566 | 1 | |
| Deletion | Chr3 | 2949877 | 2949885 | 8 | LOC_Os03g05880 |
| Deletion | Chr3 | 3694205 | 3694231 | 26 | LOC_Os03g07220 |
| Deletion | Chr3 | 7857254 | 7857266 | 12 | |
| Deletion | Chr7 | 9702884 | 9702885 | 1 | LOC_Os07g16540 |
| Deletion | Chr3 | 10581325 | 10581352 | 27 | |
| Deletion | Chr10 | 12004354 | 12004355 | 1 | |
| Deletion | Chr7 | 15348016 | 15348019 | 3 | |
| Deletion | Chr3 | 15845528 | 15845532 | 4 | |
| Deletion | Chr9 | 17458779 | 17458794 | 15 | |
| Deletion | Chr9 | 19989643 | 19989644 | 1 | |
| Deletion | Chr4 | 20738020 | 20738138 | 118 | |
| Deletion | Chr8 | 21592781 | 21592793 | 12 | |
| Deletion | Chr6 | 22110333 | 22110350 | 17 | LOC_Os06g37400 |
| Deletion | Chr9 | 22675836 | 22675844 | 8 | |
| Deletion | Chr5 | 23815405 | 23815407 | 2 | |
| Deletion | Chr1 | 25367064 | 25367073 | 9 | LOC_Os01g44230 |
| Deletion | Chr8 | 26497925 | 26498413 | 488 | |
| Deletion | Chr6 | 27762748 | 27762755 | 7 | |
| Deletion | Chr11 | 28522961 | 28522965 | 4 | LOC_Os11g47420 |
| Deletion | Chr2 | 29529411 | 29529465 | 54 | |
| Deletion | Chr2 | 33167790 | 33167831 | 41 |
Insertions: 1
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr11 | 22223380 | 22223383 | 4 |
Inversions: 3
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr12 | 463366 | 1368675 | |
| Inversion | Chr10 | 3191223 | 3449768 | LOC_Os10g06640 |
| Inversion | Chr1 | 38819618 | 40236748 | 2 |
Translocations: 2
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr12 | 12895647 | Chr4 | 29635117 | |
| Translocation | Chr8 | 19315827 | Chr4 | 6100392 | LOC_Os08g31240 |
