Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN4522-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN4522-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 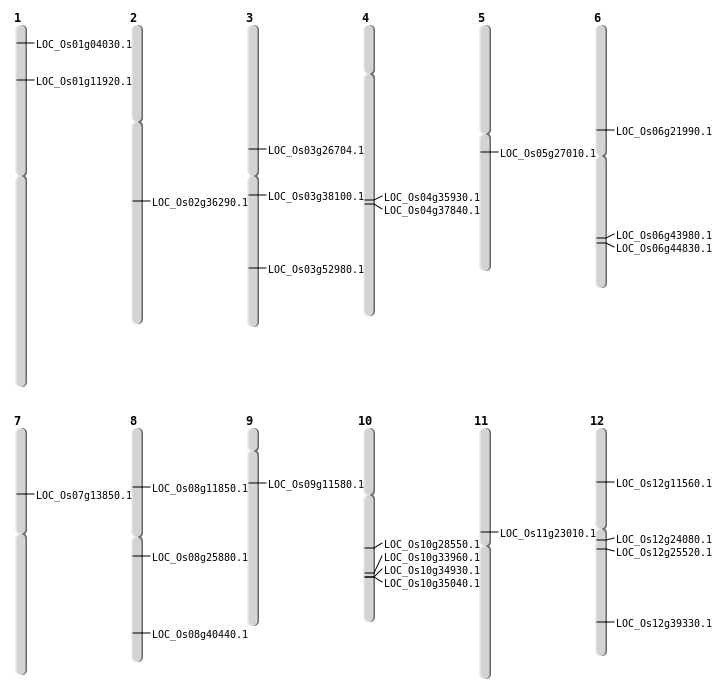
|
| Variant Information |
|---|
Single base substitutions: 66
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 20505642 | C-A | HET | ||
| SBS | Chr1 | 22078583 | C-A | HET | ||
| SBS | Chr1 | 26915073 | A-G | HET | ||
| SBS | Chr1 | 29788411 | G-T | HET | ||
| SBS | Chr1 | 35731286 | C-T | HOMO | ||
| SBS | Chr10 | 12300840 | C-T | HOMO | ||
| SBS | Chr10 | 16062726 | A-T | HET | ||
| SBS | Chr10 | 18087356 | C-T | HET | LOC_Os10g33960 | |
| SBS | Chr10 | 18641729 | G-A | HOMO | LOC_Os10g34930 | |
| SBS | Chr10 | 18700050 | C-A | HET | LOC_Os10g35040 | |
| SBS | Chr10 | 21516177 | G-C | HET | ||
| SBS | Chr10 | 9493803 | C-T | HET | ||
| SBS | Chr11 | 13233608 | G-A | HET | LOC_Os11g23010 | |
| SBS | Chr11 | 21055368 | C-T | HET | ||
| SBS | Chr11 | 25515968 | C-T | HET | ||
| SBS | Chr11 | 25516005 | A-T | HET | ||
| SBS | Chr12 | 12644223 | C-A | HET | ||
| SBS | Chr12 | 13698515 | C-A | HET | LOC_Os12g24080 | |
| SBS | Chr12 | 58493 | G-T | HET | ||
| SBS | Chr2 | 19876440 | G-T | HET | ||
| SBS | Chr2 | 24176236 | G-A | HET | ||
| SBS | Chr2 | 25023842 | C-T | HET | ||
| SBS | Chr2 | 30894087 | A-G | HET | ||
| SBS | Chr2 | 4574308 | C-A | HOMO | ||
| SBS | Chr2 | 4574309 | T-A | HOMO | ||
| SBS | Chr2 | 65470 | C-T | HET | ||
| SBS | Chr3 | 10124759 | G-A | HET | ||
| SBS | Chr3 | 17648050 | C-T | HOMO | ||
| SBS | Chr3 | 28008677 | C-T | HET | ||
| SBS | Chr3 | 30869229 | C-T | HET | ||
| SBS | Chr3 | 34533001 | T-C | HET | ||
| SBS | Chr4 | 1700315 | A-G | HET | ||
| SBS | Chr4 | 22498587 | C-T | HET | LOC_Os04g37840 | |
| SBS | Chr4 | 3180251 | G-A | HET | ||
| SBS | Chr4 | 5984193 | G-C | HET | ||
| SBS | Chr5 | 10263828 | C-T | HET | ||
| SBS | Chr5 | 15709518 | C-T | HET | LOC_Os05g27010 | |
| SBS | Chr5 | 18878050 | G-T | HET | ||
| SBS | Chr5 | 19616334 | A-T | HET | ||
| SBS | Chr5 | 19616337 | G-T | HET | ||
| SBS | Chr5 | 24601964 | G-A | HET | ||
| SBS | Chr5 | 7029727 | T-C | HET | ||
| SBS | Chr5 | 8936415 | A-G | HET | ||
| SBS | Chr6 | 13451574 | C-T | HOMO | ||
| SBS | Chr6 | 16509770 | T-C | HET | ||
| SBS | Chr6 | 17677132 | T-A | HET | ||
| SBS | Chr6 | 21359061 | G-A | HOMO | ||
| SBS | Chr6 | 3656005 | G-T | HET | ||
| SBS | Chr6 | 9806169 | T-A | HET | ||
| SBS | Chr7 | 10534262 | A-T | HOMO | ||
| SBS | Chr7 | 10534263 | A-T | HOMO | ||
| SBS | Chr7 | 24005781 | C-A | HOMO | ||
| SBS | Chr7 | 2936236 | T-A | HOMO | ||
| SBS | Chr7 | 5639640 | C-A | HOMO | ||
| SBS | Chr7 | 5990290 | G-T | HET | ||
| SBS | Chr7 | 7930300 | C-T | HOMO | LOC_Os07g13850 | |
| SBS | Chr7 | 7930301 | G-A | HOMO | ||
| SBS | Chr8 | 11386885 | A-T | HET | ||
| SBS | Chr8 | 23645612 | T-G | HET | ||
| SBS | Chr8 | 25592103 | C-T | HET | LOC_Os08g40440 | |
| SBS | Chr8 | 5312869 | G-A | HET | ||
| SBS | Chr8 | 6950254 | C-T | HOMO | LOC_Os08g11850 | |
| SBS | Chr9 | 22074791 | G-A | HET | ||
| SBS | Chr9 | 3513362 | G-A | HET | ||
| SBS | Chr9 | 492647 | T-A | HET | ||
| SBS | Chr9 | 6464308 | C-T | HET | LOC_Os09g11580 |
Deletions: 31
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr7 | 1989766 | 1989766 | 1 | |
| Deletion | Chr9 | 2196119 | 2196119 | 1 | |
| Deletion | Chr8 | 2724643 | 2724643 | 1 | |
| Deletion | Chr1 | 3833331 | 3833338 | 8 | |
| Deletion | Chr2 | 4539529 | 4539529 | 1 | |
| Deletion | Chr4 | 6205591 | 6205591 | 1 | |
| Deletion | Chr1 | 6215879 | 6215879 | 1 | |
| Deletion | Chr11 | 6469102 | 6469103 | 2 | |
| Deletion | Chr3 | 6692915 | 6692915 | 1 | |
| Deletion | Chr4 | 7071480 | 7071480 | 1 | |
| Deletion | Chr10 | 7888226 | 7888226 | 1 | |
| Deletion | Chr7 | 7930353 | 7930353 | 1 | LOC_Os07g13850 |
| Deletion | Chr10 | 8140190 | 8140211 | 22 | |
| Deletion | Chr12 | 8515215 | 8515228 | 14 | |
| Deletion | Chr11 | 9846705 | 9846705 | 1 | |
| Deletion | Chr12 | 12302108 | 12302121 | 14 | |
| Deletion | Chr2 | 14822750 | 14822761 | 12 | |
| Deletion | Chr9 | 15454288 | 15454300 | 13 | |
| Deletion | Chr5 | 16935363 | 16935366 | 4 | |
| Deletion | Chr1 | 16941512 | 16941521 | 10 | |
| Deletion | Chr5 | 17536534 | 17536534 | 1 | |
| Deletion | Chr12 | 18143944 | 18143946 | 3 | |
| Deletion | Chr7 | 21199611 | 21199611 | 1 | |
| Deletion | Chr1 | 21568325 | 21568326 | 2 | |
| Deletion | Chr1 | 21800927 | 21800931 | 5 | |
| Deletion | Chr2 | 22487980 | 22488021 | 42 | |
| Deletion | Chr5 | 22850177 | 22850177 | 1 | |
| Deletion | Chr6 | 26490497 | 26490516 | 20 | LOC_Os06g43980 |
| Deletion | Chr6 | 30409545 | 30409549 | 5 | |
| Deletion | Chr1 | 36337403 | 36337403 | 1 | |
| Deletion | Chr1 | 42765131 | 42765148 | 18 |
Insertions: 17
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr1 | 36434601 | 36434629 | 29 | |
| Insertion | Chr1 | 38787938 | 38787938 | 1 | |
| Insertion | Chr10 | 14002198 | 14002198 | 1 | |
| Insertion | Chr10 | 20304360 | 20304365 | 6 | |
| Insertion | Chr10 | 8802629 | 8802629 | 1 | |
| Insertion | Chr11 | 15578811 | 15578816 | 6 | |
| Insertion | Chr12 | 11836374 | 11836419 | 46 | |
| Insertion | Chr12 | 12666970 | 12666970 | 1 | |
| Insertion | Chr2 | 20440560 | 20440561 | 2 | |
| Insertion | Chr2 | 21913119 | 21913119 | 1 | LOC_Os02g36290 |
| Insertion | Chr4 | 29369797 | 29369798 | 2 | |
| Insertion | Chr4 | 31598752 | 31598753 | 2 | |
| Insertion | Chr5 | 18570978 | 18570978 | 1 | |
| Insertion | Chr6 | 21519232 | 21519232 | 1 | |
| Insertion | Chr8 | 17507783 | 17507825 | 43 | |
| Insertion | Chr8 | 20674425 | 20674468 | 44 | |
| Insertion | Chr9 | 18633431 | 18633432 | 2 |
Inversions: 4
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr1 | 1754460 | 2102853 | LOC_Os01g04030 |
| Inversion | Chr1 | 1754461 | 2102867 | LOC_Os01g04030 |
| Inversion | Chr9 | 8849155 | 8849418 | |
| Inversion | Chr6 | 12718778 | 12719076 | LOC_Os06g21990 |
Translocations: 3
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr8 | 15765284 | Chr3 | 15248054 | 2 |
| Translocation | Chr5 | 16732072 | Chr4 | 21907968 | LOC_Os04g35930 |
| Translocation | Chr5 | 16787806 | Chr4 | 21909499 |
