Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN4524-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN4524-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 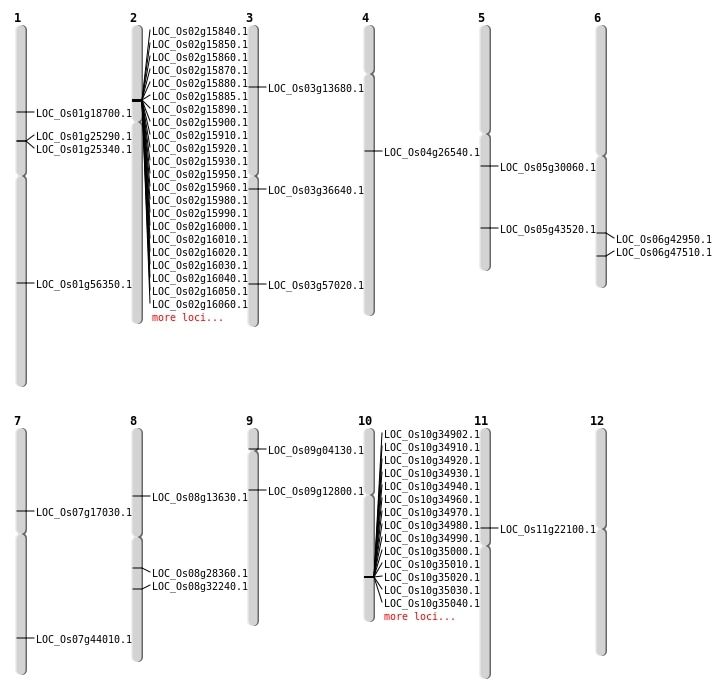
|
| Variant Information |
|---|
Single base substitutions: 36
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 33013414 | G-T | HET | ||
| SBS | Chr1 | 5185032 | G-A | HET | ||
| SBS | Chr10 | 1274939 | G-T | HET | ||
| SBS | Chr10 | 1274940 | C-A | HET | ||
| SBS | Chr10 | 17667383 | T-C | HET | ||
| SBS | Chr10 | 20280358 | G-A | HET | ||
| SBS | Chr11 | 12690201 | G-A | HET | LOC_Os11g22100 | |
| SBS | Chr11 | 16726608 | C-T | HET | ||
| SBS | Chr11 | 1942405 | T-A | HET | ||
| SBS | Chr11 | 19516589 | A-G | HET | ||
| SBS | Chr11 | 27257793 | C-G | HOMO | ||
| SBS | Chr12 | 10280209 | G-T | HET | ||
| SBS | Chr12 | 4452991 | A-G | HET | ||
| SBS | Chr2 | 10241212 | A-G | HET | ||
| SBS | Chr2 | 21092324 | A-G | HET | ||
| SBS | Chr2 | 26027789 | C-G | HET | ||
| SBS | Chr2 | 6634294 | G-A | HET | ||
| SBS | Chr2 | 6634295 | C-T | HET | ||
| SBS | Chr3 | 28661286 | C-T | HET | ||
| SBS | Chr3 | 28940889 | C-T | HET | ||
| SBS | Chr3 | 28984926 | A-G | HOMO | ||
| SBS | Chr3 | 32485223 | T-C | HET | LOC_Os03g57020 | |
| SBS | Chr4 | 16897980 | G-A | HOMO | ||
| SBS | Chr4 | 21638654 | T-A | HET | ||
| SBS | Chr5 | 17390598 | T-G | HET | LOC_Os05g30060 | |
| SBS | Chr5 | 22398785 | C-A | HET | LOC_Os05g38190 | |
| SBS | Chr5 | 23039474 | G-A | HET | ||
| SBS | Chr5 | 26374021 | G-A | HET | ||
| SBS | Chr5 | 786530 | T-C | HET | ||
| SBS | Chr6 | 28782381 | G-C | HOMO | ||
| SBS | Chr8 | 18617394 | T-G | HET | ||
| SBS | Chr8 | 19977059 | T-A | HET | LOC_Os08g32240 | |
| SBS | Chr8 | 23983605 | G-A | HET | ||
| SBS | Chr9 | 10773994 | A-C | HOMO | ||
| SBS | Chr9 | 20118377 | A-C | HET | ||
| SBS | Chr9 | 7350790 | G-T | HOMO | LOC_Os09g12800 |
Deletions: 32
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr3 | 587433 | 587433 | 1 | |
| Deletion | Chr8 | 781081 | 781081 | 1 | |
| Deletion | Chr7 | 2928704 | 2928704 | 1 | |
| Deletion | Chr2 | 4468463 | 4468463 | 1 | |
| Deletion | Chr8 | 4911835 | 4911835 | 1 | |
| Deletion | Chr12 | 5640680 | 5640680 | 1 | |
| Deletion | Chr3 | 6674069 | 6674069 | 1 | |
| Deletion | Chr2 | 8951001 | 9263000 | 312000 | 43 |
| Deletion | Chr4 | 9821550 | 9821554 | 5 | |
| Deletion | Chr7 | 10027937 | 10027937 | 1 | LOC_Os07g17030 |
| Deletion | Chr8 | 11012203 | 11012204 | 2 | |
| Deletion | Chr11 | 11996780 | 11996780 | 1 | |
| Deletion | Chr8 | 13015583 | 13015583 | 1 | |
| Deletion | Chr2 | 13090647 | 13090647 | 1 | |
| Deletion | Chr11 | 15738570 | 15738575 | 6 | |
| Deletion | Chr6 | 16047394 | 16047461 | 68 | |
| Deletion | Chr4 | 16683902 | 16683911 | 10 | |
| Deletion | Chr8 | 17305143 | 17305155 | 13 | LOC_Os08g28360 |
| Deletion | Chr10 | 18630001 | 18742000 | 112000 | 21 |
| Deletion | Chr10 | 18881001 | 18910000 | 29000 | 8 |
| Deletion | Chr9 | 20176168 | 20176208 | 41 | |
| Deletion | Chr10 | 20402589 | 20402589 | 1 | |
| Deletion | Chr2 | 21079701 | 21079701 | 1 | |
| Deletion | Chr5 | 21863696 | 21863696 | 1 | |
| Deletion | Chr4 | 21995775 | 21995779 | 5 | |
| Deletion | Chr5 | 25298427 | 25298432 | 6 | LOC_Os05g43520 |
| Deletion | Chr11 | 25530968 | 25530968 | 1 | |
| Deletion | Chr6 | 25806055 | 25806055 | 1 | LOC_Os06g42950 |
| Deletion | Chr7 | 26308311 | 26308311 | 1 | LOC_Os07g44010 |
| Deletion | Chr1 | 26379274 | 26379274 | 1 | |
| Deletion | Chr5 | 29941679 | 29941679 | 1 | |
| Deletion | Chr1 | 36614284 | 36614287 | 4 |
Insertions: 20
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr1 | 2797500 | 2797616 | 117 | |
| Insertion | Chr1 | 4185094 | 4185094 | 1 | |
| Insertion | Chr10 | 20977113 | 20977113 | 1 | |
| Insertion | Chr10 | 2477980 | 2478016 | 37 | |
| Insertion | Chr11 | 790137 | 790137 | 1 | |
| Insertion | Chr11 | 7920716 | 7920717 | 2 | |
| Insertion | Chr2 | 12409836 | 12409844 | 9 | |
| Insertion | Chr2 | 13661073 | 13661073 | 1 | |
| Insertion | Chr2 | 24066660 | 24066788 | 129 | |
| Insertion | Chr2 | 24448708 | 24448708 | 1 | |
| Insertion | Chr3 | 23141900 | 23141905 | 6 | |
| Insertion | Chr4 | 26456788 | 26456790 | 3 | |
| Insertion | Chr4 | 35486289 | 35486289 | 1 | |
| Insertion | Chr5 | 17221409 | 17221525 | 117 | |
| Insertion | Chr5 | 6123378 | 6123379 | 2 | |
| Insertion | Chr5 | 6127134 | 6127134 | 1 | |
| Insertion | Chr6 | 28763937 | 28763939 | 3 | LOC_Os06g47510 |
| Insertion | Chr7 | 21071787 | 21071881 | 95 | |
| Insertion | Chr7 | 26337083 | 26337189 | 107 | |
| Insertion | Chr8 | 10992755 | 10992755 | 1 |
Inversions: 5
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr8 | 8106963 | 8107116 | LOC_Os08g13630 |
| Inversion | Chr2 | 11343472 | 11343581 | |
| Inversion | Chr10 | 18632408 | 18880917 | |
| Inversion | Chr12 | 21772582 | 21772698 | |
| Inversion | Chr1 | 32475935 | 32476112 | LOC_Os01g56350 |
Translocations: 3
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr10 | 20767587 | Chr1 | 30620216 | |
| Translocation | Chr10 | 20767590 | Chr1 | 30620220 | |
| Translocation | Chr5 | 27825501 | Chr2 | 11342526 |
