Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN4529-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN4529-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 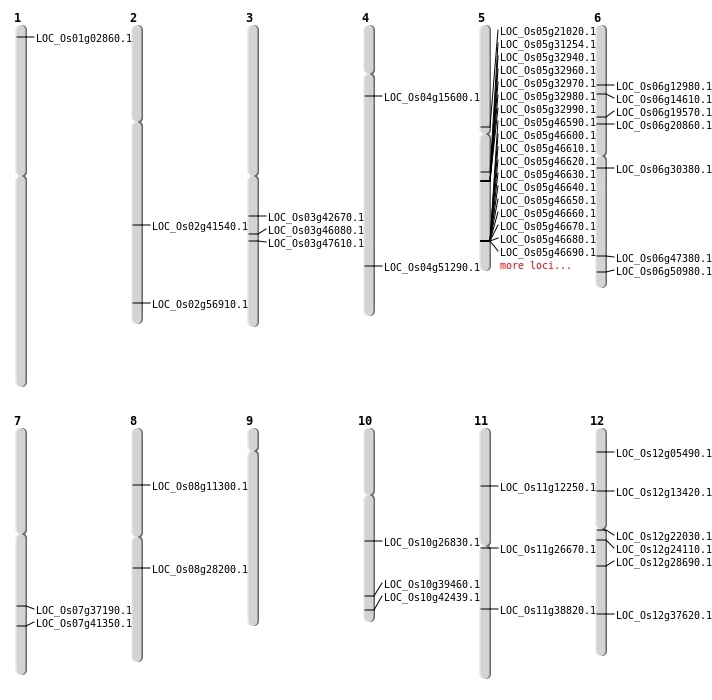
|
| Variant Information |
|---|
Single base substitutions: 106
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 1003187 | G-A | HET | LOC_Os01g02860 | |
| SBS | Chr1 | 12920229 | A-G | HET | ||
| SBS | Chr1 | 21618032 | C-T | HET | ||
| SBS | Chr1 | 23232215 | G-A | HOMO | ||
| SBS | Chr1 | 23232216 | T-C | HOMO | ||
| SBS | Chr1 | 23232217 | G-T | HOMO | ||
| SBS | Chr1 | 24303197 | T-C | HET | ||
| SBS | Chr1 | 26453056 | T-A | HET | ||
| SBS | Chr1 | 29746244 | C-A | HET | ||
| SBS | Chr1 | 31714076 | A-T | HET | ||
| SBS | Chr1 | 33901343 | T-A | HET | ||
| SBS | Chr1 | 37634852 | T-C | HET | ||
| SBS | Chr10 | 15260002 | C-A | HET | ||
| SBS | Chr10 | 2069652 | C-A | HET | ||
| SBS | Chr10 | 22287828 | C-T | HET | ||
| SBS | Chr10 | 22868888 | G-T | HET | LOC_Os10g42439 | |
| SBS | Chr11 | 12993522 | A-G | HET | ||
| SBS | Chr11 | 15278725 | G-T | HET | LOC_Os11g26670 | |
| SBS | Chr11 | 23091428 | C-A | HET | LOC_Os11g38820 | |
| SBS | Chr11 | 484957 | C-A | HET | ||
| SBS | Chr11 | 7369897 | C-A | HET | ||
| SBS | Chr11 | 8680663 | G-A | HET | ||
| SBS | Chr12 | 12375029 | T-C | HET | ||
| SBS | Chr12 | 12394335 | G-T | HET | LOC_Os12g22030 | |
| SBS | Chr12 | 13716881 | G-A | HET | LOC_Os12g24110 | |
| SBS | Chr12 | 15359834 | G-C | HET | ||
| SBS | Chr12 | 15834728 | T-C | HET | ||
| SBS | Chr12 | 16653531 | A-T | HET | ||
| SBS | Chr12 | 17103260 | T-C | HET | ||
| SBS | Chr12 | 22334512 | C-G | HET | ||
| SBS | Chr12 | 23100663 | C-A | HET | LOC_Os12g37620 | |
| SBS | Chr12 | 2511046 | G-A | HET | LOC_Os12g05490 | |
| SBS | Chr12 | 25417186 | T-A | HOMO | ||
| SBS | Chr12 | 25417188 | G-A | HOMO | ||
| SBS | Chr12 | 26689069 | C-A | HET | ||
| SBS | Chr12 | 4440277 | G-A | HET | ||
| SBS | Chr12 | 6592268 | T-C | HET | ||
| SBS | Chr2 | 12114032 | A-G | HET | ||
| SBS | Chr2 | 15942022 | C-T | HET | ||
| SBS | Chr2 | 17840011 | G-T | HET | ||
| SBS | Chr2 | 19481002 | C-T | HET | ||
| SBS | Chr2 | 23028724 | T-C | HOMO | ||
| SBS | Chr2 | 24911688 | G-C | HET | LOC_Os02g41540 | |
| SBS | Chr2 | 34884946 | A-G | HET | LOC_Os02g56910 | |
| SBS | Chr3 | 14840994 | T-A | HET | ||
| SBS | Chr3 | 15620590 | T-C | HET | ||
| SBS | Chr3 | 1701456 | G-A | HET | ||
| SBS | Chr3 | 18818890 | C-G | HOMO | ||
| SBS | Chr3 | 22518048 | G-A | HET | ||
| SBS | Chr3 | 2593373 | C-A | HOMO | ||
| SBS | Chr3 | 26058821 | G-A | HET | LOC_Os03g46080 | |
| SBS | Chr3 | 26954686 | G-T | HET | LOC_Os03g47610 | |
| SBS | Chr3 | 32123475 | T-A | HET | ||
| SBS | Chr3 | 4845032 | C-T | HOMO | ||
| SBS | Chr3 | 6920134 | T-A | HET | ||
| SBS | Chr3 | 6920135 | T-A | HET | ||
| SBS | Chr3 | 6920136 | G-A | HET | ||
| SBS | Chr3 | 7969678 | A-G | HET | ||
| SBS | Chr3 | 8806433 | C-A | HET | ||
| SBS | Chr4 | 20002422 | G-A | HOMO | ||
| SBS | Chr4 | 2288208 | G-A | HET | ||
| SBS | Chr4 | 23826620 | C-A | HET | ||
| SBS | Chr4 | 35034575 | T-C | HET | ||
| SBS | Chr4 | 8485537 | C-A | HET | LOC_Os04g15600 | |
| SBS | Chr5 | 16827775 | A-G | HET | ||
| SBS | Chr5 | 18180580 | T-A | HET | LOC_Os05g31254 | |
| SBS | Chr5 | 18180581 | G-A | HET | LOC_Os05g31254 | |
| SBS | Chr5 | 18788450 | G-A | HET | ||
| SBS | Chr5 | 19151228 | A-C | HET | ||
| SBS | Chr5 | 19295734 | G-A | HET | LOC_Os05g32940 | |
| SBS | Chr5 | 19361825 | A-T | HET | ||
| SBS | Chr5 | 23073199 | C-A | HET | ||
| SBS | Chr5 | 26506731 | G-T | HET | ||
| SBS | Chr5 | 3689735 | T-C | HET | ||
| SBS | Chr5 | 702039 | C-T | HET | ||
| SBS | Chr6 | 11931589 | A-G | HET | ||
| SBS | Chr6 | 12132365 | C-T | HET | ||
| SBS | Chr6 | 13629638 | G-T | HET | ||
| SBS | Chr6 | 17553015 | G-T | HET | LOC_Os06g30380 | |
| SBS | Chr6 | 22192750 | T-A | HET | ||
| SBS | Chr6 | 2387111 | T-G | HET | ||
| SBS | Chr6 | 5597843 | G-A | HET | ||
| SBS | Chr6 | 8223720 | G-A | HET | LOC_Os06g14610 | |
| SBS | Chr6 | 9819896 | G-A | HET | ||
| SBS | Chr7 | 10594948 | G-T | HET | ||
| SBS | Chr7 | 12622262 | G-T | HOMO | ||
| SBS | Chr7 | 22282377 | A-T | HET | LOC_Os07g37190 | |
| SBS | Chr7 | 2234828 | T-A | HET | ||
| SBS | Chr7 | 23249433 | A-G | HET | ||
| SBS | Chr7 | 24768843 | C-T | HET | LOC_Os07g41350 | |
| SBS | Chr7 | 382889 | A-G | HET | ||
| SBS | Chr7 | 7296632 | C-T | HET | ||
| SBS | Chr7 | 9419866 | A-G | HET | ||
| SBS | Chr8 | 12894398 | A-G | HET | ||
| SBS | Chr8 | 14121747 | C-A | HET | ||
| SBS | Chr8 | 15971212 | C-T | HET | ||
| SBS | Chr9 | 11059680 | A-T | HOMO | ||
| SBS | Chr9 | 12966780 | A-C | HOMO | ||
| SBS | Chr9 | 13623692 | G-T | HOMO | ||
| SBS | Chr9 | 16527067 | A-G | HET | ||
| SBS | Chr9 | 16707443 | C-T | HET | ||
| SBS | Chr9 | 2047937 | C-T | HET | ||
| SBS | Chr9 | 21324014 | C-A | HET | ||
| SBS | Chr9 | 4410657 | C-T | HET | ||
| SBS | Chr9 | 5781687 | C-A | HET | ||
| SBS | Chr9 | 8938422 | C-T | HET |
Deletions: 27
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr11 | 2178755 | 2178755 | 1 | |
| Deletion | Chr4 | 3331851 | 3331851 | 1 | |
| Deletion | Chr6 | 4622010 | 4622010 | 1 | |
| Deletion | Chr8 | 6648590 | 6648605 | 16 | |
| Deletion | Chr11 | 6843061 | 6843238 | 178 | LOC_Os11g12250 |
| Deletion | Chr12 | 7507217 | 7507226 | 10 | LOC_Os12g13420 |
| Deletion | Chr9 | 7836381 | 7836381 | 1 | |
| Deletion | Chr6 | 9840281 | 9840281 | 1 | |
| Deletion | Chr12 | 10793249 | 10793251 | 3 | |
| Deletion | Chr10 | 12705045 | 12705045 | 1 | |
| Deletion | Chr1 | 13729515 | 13729619 | 105 | |
| Deletion | Chr12 | 15195240 | 15195309 | 70 | |
| Deletion | Chr4 | 15219285 | 15219371 | 87 | |
| Deletion | Chr12 | 16952123 | 16952181 | 59 | LOC_Os12g28690 |
| Deletion | Chr5 | 17756958 | 17756958 | 1 | |
| Deletion | Chr5 | 19313001 | 19337000 | 24000 | 4 |
| Deletion | Chr4 | 19821921 | 19821921 | 1 | |
| Deletion | Chr10 | 21058496 | 21058529 | 34 | LOC_Os10g39460 |
| Deletion | Chr12 | 21920980 | 21920981 | 2 | |
| Deletion | Chr12 | 23024348 | 23024369 | 22 | |
| Deletion | Chr1 | 23236305 | 23236307 | 3 | |
| Deletion | Chr2 | 23617523 | 23617523 | 1 | |
| Deletion | Chr12 | 26811495 | 26811495 | 1 | |
| Deletion | Chr5 | 26972001 | 27128000 | 156000 | 27 |
| Deletion | Chr6 | 28718710 | 28718715 | 6 | LOC_Os06g47380 |
| Deletion | Chr6 | 29012517 | 29012610 | 94 | |
| Deletion | Chr4 | 29698443 | 29698456 | 14 |
Insertions: 28
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr1 | 12538303 | 12538303 | 1 | |
| Insertion | Chr1 | 19991226 | 19991228 | 3 | |
| Insertion | Chr1 | 29661307 | 29661307 | 1 | |
| Insertion | Chr10 | 3061973 | 3061973 | 1 | |
| Insertion | Chr10 | 786700 | 786700 | 1 | |
| Insertion | Chr11 | 2163968 | 2163968 | 1 | |
| Insertion | Chr11 | 23031920 | 23031961 | 42 | |
| Insertion | Chr11 | 24993502 | 24993502 | 1 | |
| Insertion | Chr11 | 4262535 | 4262557 | 23 | |
| Insertion | Chr12 | 11016854 | 11016854 | 1 | |
| Insertion | Chr12 | 12616516 | 12616516 | 1 | |
| Insertion | Chr2 | 32654577 | 32654593 | 17 | |
| Insertion | Chr2 | 33662626 | 33662626 | 1 | |
| Insertion | Chr2 | 5542151 | 5542152 | 2 | |
| Insertion | Chr2 | 7458562 | 7458566 | 5 | |
| Insertion | Chr3 | 15055195 | 15055195 | 1 | |
| Insertion | Chr3 | 915058 | 915058 | 1 | |
| Insertion | Chr4 | 11634229 | 11634350 | 122 | |
| Insertion | Chr4 | 17209076 | 17209076 | 1 | |
| Insertion | Chr4 | 26648739 | 26648812 | 74 | |
| Insertion | Chr5 | 12367664 | 12367666 | 3 | LOC_Os05g21020 |
| Insertion | Chr6 | 11144460 | 11144484 | 25 | LOC_Os06g19570 |
| Insertion | Chr6 | 31125907 | 31125907 | 1 | |
| Insertion | Chr6 | 8045661 | 8045661 | 1 | |
| Insertion | Chr7 | 18061748 | 18061748 | 1 | |
| Insertion | Chr7 | 22481083 | 22481083 | 1 | |
| Insertion | Chr7 | 686241 | 686241 | 1 | |
| Insertion | Chr8 | 9587328 | 9587328 | 1 |
Inversions: 7
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr9 | 6707237 | 6707500 | |
| Inversion | Chr6 | 7102747 | 7102948 | LOC_Os06g12980 |
| Inversion | Chr7 | 8146825 | 8146991 | |
| Inversion | Chr8 | 8981784 | 8981873 | |
| Inversion | Chr10 | 9926236 | 9926374 | |
| Inversion | Chr4 | 13008549 | 13008738 | |
| Inversion | Chr7 | 16020230 | 16020473 |
No Translocation
