Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN4533-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN4533-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 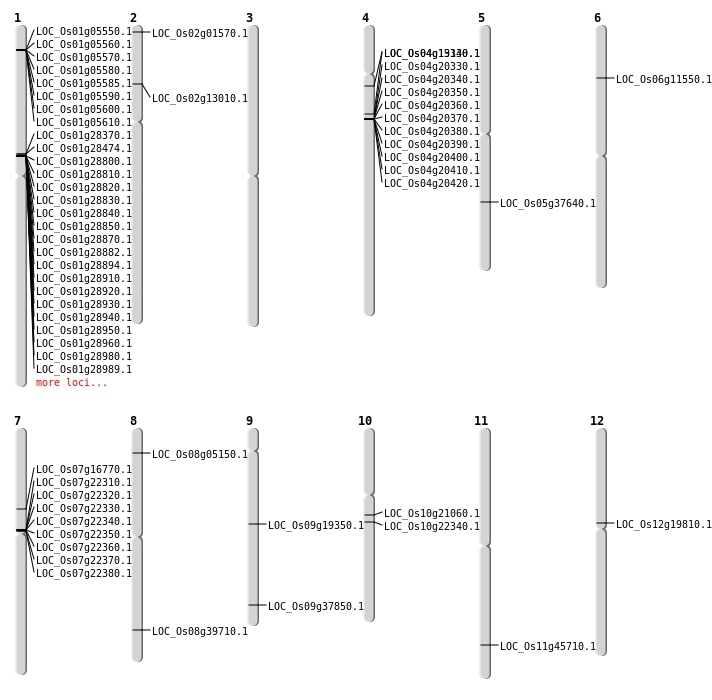
|
| Variant Information |
|---|
Single base substitutions: 54
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 12513187 | G-A | HOMO | ||
| SBS | Chr1 | 14978694 | T-C | HET | ||
| SBS | Chr1 | 21428496 | C-G | HOMO | ||
| SBS | Chr1 | 34437924 | C-T | HET | ||
| SBS | Chr1 | 36587342 | C-T | HET | ||
| SBS | Chr1 | 7124718 | T-A | HET | ||
| SBS | Chr10 | 11548680 | G-A | HET | LOC_Os10g22340 | |
| SBS | Chr11 | 21317837 | T-C | HOMO | ||
| SBS | Chr12 | 11196580 | T-A | HOMO | ||
| SBS | Chr12 | 11558934 | T-A | HET | LOC_Os12g19810 | |
| SBS | Chr12 | 12480421 | T-C | HET | ||
| SBS | Chr12 | 12639645 | C-T | HOMO | ||
| SBS | Chr12 | 14763429 | G-T | HET | ||
| SBS | Chr12 | 23414331 | A-T | HET | ||
| SBS | Chr12 | 24209673 | C-T | HET | ||
| SBS | Chr2 | 17926549 | A-T | HET | ||
| SBS | Chr2 | 18014838 | T-C | HET | ||
| SBS | Chr2 | 19548077 | T-C | HET | ||
| SBS | Chr2 | 30853872 | G-A | HOMO | ||
| SBS | Chr2 | 9619182 | T-A | HET | ||
| SBS | Chr3 | 10333040 | C-A | HET | ||
| SBS | Chr3 | 2081187 | G-T | HOMO | ||
| SBS | Chr3 | 21779910 | A-G | HET | ||
| SBS | Chr3 | 22748306 | T-C | HET | ||
| SBS | Chr3 | 32639273 | C-T | HET | ||
| SBS | Chr4 | 10168300 | T-G | HET | ||
| SBS | Chr4 | 10753898 | G-A | HET | LOC_Os04g19330 | |
| SBS | Chr4 | 10880931 | G-A | HET | ||
| SBS | Chr4 | 24589695 | C-T | HET | ||
| SBS | Chr4 | 27712770 | G-C | HET | ||
| SBS | Chr4 | 3113788 | A-G | HET | ||
| SBS | Chr4 | 33090681 | G-T | HET | ||
| SBS | Chr4 | 5192761 | T-A | HET | ||
| SBS | Chr5 | 15305393 | G-A | HOMO | ||
| SBS | Chr5 | 22029609 | A-T | HOMO | LOC_Os05g37640 | |
| SBS | Chr5 | 24772742 | T-C | HET | ||
| SBS | Chr5 | 3389125 | C-T | HET | ||
| SBS | Chr6 | 15251095 | C-T | HET | ||
| SBS | Chr6 | 17638682 | C-T | HET | ||
| SBS | Chr6 | 21883021 | C-A | HET | ||
| SBS | Chr6 | 28514771 | G-A | HET | ||
| SBS | Chr6 | 30108411 | G-T | HET | ||
| SBS | Chr7 | 17622848 | G-C | HET | ||
| SBS | Chr7 | 21475367 | A-G | HOMO | ||
| SBS | Chr7 | 28872022 | G-A | HET | ||
| SBS | Chr7 | 4336724 | G-C | HET | ||
| SBS | Chr7 | 9843828 | C-T | HET | LOC_Os07g16770 | |
| SBS | Chr8 | 18151300 | G-T | HET | ||
| SBS | Chr8 | 20772386 | A-T | HOMO | ||
| SBS | Chr8 | 24889981 | C-G | HET | ||
| SBS | Chr8 | 2670285 | T-A | HOMO | LOC_Os08g05150 | |
| SBS | Chr9 | 11578720 | T-C | HOMO | LOC_Os09g19350 | |
| SBS | Chr9 | 14341397 | G-A | HET | ||
| SBS | Chr9 | 4366849 | G-A | HOMO |
Deletions: 35
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr2 | 1243843 | 1243843 | 1 | |
| Deletion | Chr12 | 2049860 | 2049860 | 1 | |
| Deletion | Chr1 | 2646001 | 2675000 | 29000 | 8 |
| Deletion | Chr12 | 3723035 | 3723035 | 1 | |
| Deletion | Chr7 | 5041440 | 5041440 | 1 | |
| Deletion | Chr1 | 5341491 | 5341491 | 1 | |
| Deletion | Chr9 | 5451168 | 5451170 | 3 | |
| Deletion | Chr5 | 5538904 | 5538905 | 2 | |
| Deletion | Chr6 | 6122990 | 6122996 | 7 | LOC_Os06g11550 |
| Deletion | Chr2 | 6894596 | 6894596 | 1 | LOC_Os02g13010 |
| Deletion | Chr2 | 7398665 | 7398665 | 1 | |
| Deletion | Chr12 | 10293028 | 10293028 | 1 | |
| Deletion | Chr4 | 11233766 | 11233767 | 2 | |
| Deletion | Chr4 | 11377001 | 11422000 | 45000 | 10 |
| Deletion | Chr2 | 11823998 | 11824010 | 13 | |
| Deletion | Chr7 | 12507001 | 12564000 | 57000 | 8 |
| Deletion | Chr12 | 13441978 | 13441979 | 2 | |
| Deletion | Chr3 | 13484351 | 13484351 | 1 | |
| Deletion | Chr1 | 15889001 | 15899000 | 10000 | LOC_Os01g28370 |
| Deletion | Chr1 | 16135001 | 16269000 | 134000 | 24 |
| Deletion | Chr1 | 16274001 | 16309000 | 35000 | 6 |
| Deletion | Chr1 | 16312001 | 16555000 | 243000 | 33 |
| Deletion | Chr11 | 16494988 | 16494990 | 3 | |
| Deletion | Chr11 | 16518111 | 16518111 | 1 | |
| Deletion | Chr1 | 16787296 | 16787296 | 1 | |
| Deletion | Chr7 | 17153914 | 17153915 | 2 | |
| Deletion | Chr9 | 17991083 | 17991083 | 1 | |
| Deletion | Chr9 | 18587942 | 18587942 | 1 | |
| Deletion | Chr5 | 21307788 | 21307794 | 7 | |
| Deletion | Chr2 | 21696318 | 21696318 | 1 | |
| Deletion | Chr5 | 22344879 | 22344884 | 6 | |
| Deletion | Chr8 | 25139768 | 25139768 | 1 | LOC_Os08g39710 |
| Deletion | Chr4 | 27274503 | 27274514 | 12 | |
| Deletion | Chr4 | 31706754 | 31706756 | 3 | |
| Deletion | Chr2 | 35122004 | 35122004 | 1 |
Insertions: 20
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr1 | 10179066 | 10179157 | 92 | |
| Insertion | Chr10 | 21082481 | 21082531 | 51 | LOC_Os10g39480 |
| Insertion | Chr10 | 21689157 | 21689158 | 2 | |
| Insertion | Chr11 | 13435197 | 13435238 | 42 | |
| Insertion | Chr12 | 7112357 | 7112385 | 29 | |
| Insertion | Chr3 | 1056609 | 1056610 | 2 | |
| Insertion | Chr3 | 14225598 | 14225599 | 2 | |
| Insertion | Chr3 | 34863825 | 34863825 | 1 | |
| Insertion | Chr4 | 21773632 | 21773632 | 1 | |
| Insertion | Chr5 | 10007414 | 10007414 | 1 | |
| Insertion | Chr5 | 1027174 | 1027177 | 4 | |
| Insertion | Chr5 | 15788763 | 15788879 | 117 | |
| Insertion | Chr5 | 25552582 | 25552582 | 1 | |
| Insertion | Chr6 | 11423230 | 11423231 | 2 | |
| Insertion | Chr8 | 14911622 | 14911628 | 7 | |
| Insertion | Chr8 | 17952539 | 17952544 | 6 | |
| Insertion | Chr9 | 1133649 | 1133746 | 98 | |
| Insertion | Chr9 | 16279523 | 16279523 | 1 | |
| Insertion | Chr9 | 5579140 | 5579184 | 45 | LOC_Os09g10240 |
| Insertion | Chr9 | 8846950 | 8846991 | 42 |
Inversions: 5
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr1 | 15178128 | 15888315 | |
| Inversion | Chr1 | 15899369 | 16560598 | |
| Inversion | Chr1 | 15948857 | 16135399 | LOC_Os01g28474 |
| Inversion | Chr6 | 18123431 | 18900965 | |
| Inversion | Chr1 | 40156043 | 40156273 |
Translocations: 3
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr12 | 18488140 | Chr11 | 18175803 | |
| Translocation | Chr9 | 21823899 | Chr1 | 2675361 | 2 |
| Translocation | Chr11 | 27652627 | Chr7 | 15855915 | LOC_Os11g45710 |
