Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN4561-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN4561-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 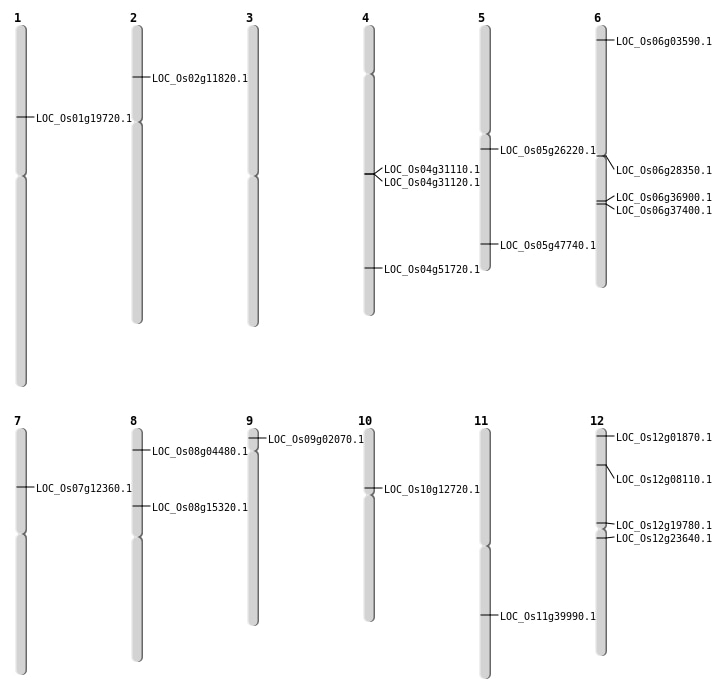
|
| Variant Information |
|---|
Single base substitutions: 59
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 12278825 | G-A | HET | ||
| SBS | Chr1 | 20197677 | G-T | HOMO | ||
| SBS | Chr1 | 28461367 | C-T | HET | ||
| SBS | Chr1 | 30301345 | T-A | HET | ||
| SBS | Chr1 | 33679758 | C-T | HET | ||
| SBS | Chr1 | 36810276 | G-A | HET | ||
| SBS | Chr1 | 40394875 | C-T | HOMO | ||
| SBS | Chr10 | 13655424 | G-A | HET | ||
| SBS | Chr10 | 15748238 | C-T | HET | ||
| SBS | Chr10 | 5503953 | C-T | HET | ||
| SBS | Chr10 | 7098066 | T-A | HET | LOC_Os10g12720 | |
| SBS | Chr11 | 13180002 | G-A | HET | ||
| SBS | Chr11 | 13267805 | G-A | HET | ||
| SBS | Chr11 | 14509577 | T-A | HET | ||
| SBS | Chr11 | 15669780 | G-T | HET | ||
| SBS | Chr11 | 16772261 | G-A | HET | ||
| SBS | Chr11 | 21593029 | G-A | HET | ||
| SBS | Chr11 | 23507976 | G-A | HET | ||
| SBS | Chr11 | 23695072 | G-A | HET | ||
| SBS | Chr11 | 23846733 | A-T | HOMO | LOC_Os11g39990 | |
| SBS | Chr12 | 10663189 | T-A | HET | ||
| SBS | Chr12 | 10663190 | G-A | HET | ||
| SBS | Chr12 | 11529170 | G-T | HET | LOC_Os12g19780 | |
| SBS | Chr12 | 13421411 | T-C | HET | LOC_Os12g23640 | |
| SBS | Chr12 | 17775761 | C-T | HET | ||
| SBS | Chr12 | 26410627 | C-T | HOMO | ||
| SBS | Chr12 | 2857450 | G-A | HET | ||
| SBS | Chr12 | 9638029 | C-G | HET | ||
| SBS | Chr2 | 12209738 | G-A | HET | ||
| SBS | Chr2 | 6115033 | C-T | HET | LOC_Os02g11820 | |
| SBS | Chr2 | 6297736 | C-T | HOMO | ||
| SBS | Chr2 | 7056312 | G-A | HOMO | ||
| SBS | Chr2 | 7497432 | T-A | HET | ||
| SBS | Chr3 | 16338644 | C-T | HET | ||
| SBS | Chr3 | 20831124 | G-A | HET | ||
| SBS | Chr3 | 33744877 | T-C | HET | ||
| SBS | Chr3 | 35953042 | A-T | HET | ||
| SBS | Chr4 | 24429411 | C-T | HET | ||
| SBS | Chr4 | 6050111 | C-T | HET | ||
| SBS | Chr5 | 21003379 | C-T | HET | ||
| SBS | Chr5 | 22668657 | G-A | HET | ||
| SBS | Chr5 | 5845925 | G-A | HET | ||
| SBS | Chr5 | 8707964 | G-A | HET | ||
| SBS | Chr6 | 11350405 | G-A | HET | ||
| SBS | Chr6 | 17364850 | G-A | HET | ||
| SBS | Chr6 | 21741964 | C-T | HET | LOC_Os06g36900 | |
| SBS | Chr6 | 24377401 | G-A | HET | ||
| SBS | Chr6 | 25037904 | G-A | HET | ||
| SBS | Chr6 | 25037905 | C-A | HET | ||
| SBS | Chr6 | 25446216 | C-T | HET | ||
| SBS | Chr7 | 7606536 | T-C | HET | ||
| SBS | Chr7 | 7805061 | A-T | HET | ||
| SBS | Chr8 | 2203512 | T-G | HOMO | LOC_Os08g04480 | |
| SBS | Chr8 | 27746761 | C-T | HOMO | ||
| SBS | Chr8 | 6574115 | C-T | HET | ||
| SBS | Chr8 | 8402236 | G-A | HET | ||
| SBS | Chr9 | 16652660 | A-T | HET | ||
| SBS | Chr9 | 2208603 | T-C | HOMO | ||
| SBS | Chr9 | 4818674 | C-T | HET |
Deletions: 35
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr9 | 106498 | 106498 | 1 | |
| Deletion | Chr6 | 525797 | 525797 | 1 | |
| Deletion | Chr4 | 637731 | 637751 | 21 | |
| Deletion | Chr6 | 1388086 | 1388379 | 294 | LOC_Os06g03590 |
| Deletion | Chr1 | 2338688 | 2338688 | 1 | |
| Deletion | Chr6 | 3906926 | 3906928 | 3 | |
| Deletion | Chr5 | 3921571 | 3921571 | 1 | |
| Deletion | Chr12 | 4133019 | 4133030 | 12 | LOC_Os12g08110 |
| Deletion | Chr4 | 6015909 | 6015928 | 20 | |
| Deletion | Chr7 | 7016089 | 7016132 | 44 | LOC_Os07g12360 |
| Deletion | Chr10 | 8158542 | 8158703 | 162 | |
| Deletion | Chr8 | 8183862 | 8183862 | 1 | |
| Deletion | Chr5 | 8712003 | 8712012 | 10 | |
| Deletion | Chr8 | 9313942 | 9313945 | 4 | LOC_Os08g15320 |
| Deletion | Chr3 | 10761809 | 10761809 | 1 | |
| Deletion | Chr3 | 11019141 | 11019141 | 1 | |
| Deletion | Chr1 | 11184213 | 11184216 | 4 | LOC_Os01g19720 |
| Deletion | Chr8 | 12482792 | 12482854 | 63 | |
| Deletion | Chr12 | 13147524 | 13147524 | 1 | |
| Deletion | Chr5 | 14255321 | 14255324 | 4 | |
| Deletion | Chr8 | 14903787 | 14903787 | 1 | |
| Deletion | Chr6 | 16127602 | 16127602 | 1 | LOC_Os06g28350 |
| Deletion | Chr6 | 16243537 | 16243537 | 1 | |
| Deletion | Chr10 | 16813487 | 16813492 | 6 | |
| Deletion | Chr7 | 18316285 | 18316285 | 1 | |
| Deletion | Chr4 | 18599001 | 18614000 | 15000 | 2 |
| Deletion | Chr2 | 19604251 | 19604256 | 6 | |
| Deletion | Chr10 | 19652952 | 19652952 | 1 | |
| Deletion | Chr12 | 20497803 | 20497844 | 42 | |
| Deletion | Chr9 | 20822442 | 20822442 | 1 | |
| Deletion | Chr7 | 22838582 | 22838582 | 1 | |
| Deletion | Chr12 | 23652369 | 23652369 | 1 | |
| Deletion | Chr1 | 26061891 | 26061891 | 1 | |
| Deletion | Chr4 | 30649682 | 30649682 | 1 | LOC_Os04g51720 |
| Deletion | Chr1 | 42238051 | 42238067 | 17 |
Insertions: 21
Inversions: 2
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr4 | 11504148 | 11504332 | |
| Inversion | Chr12 | 23189899 | 23190072 |
No Translocation
