Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN4566-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN4566-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 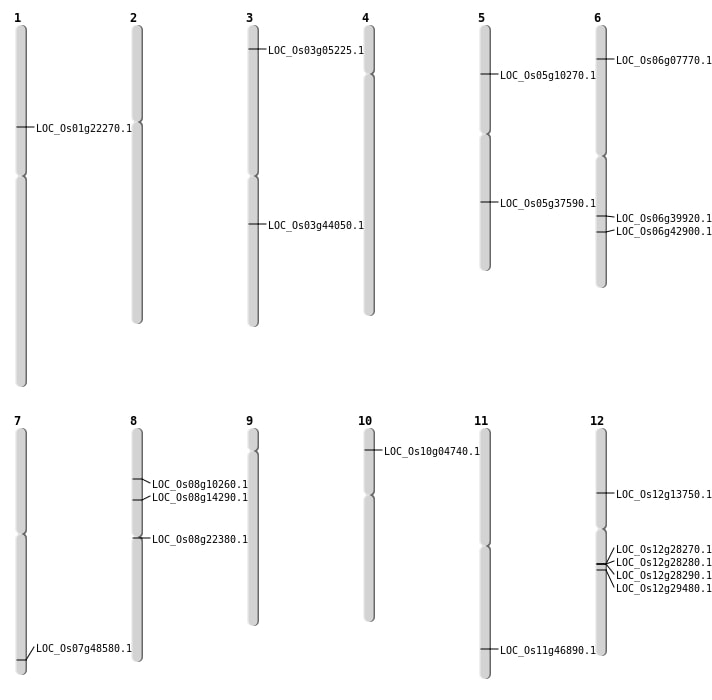
|
| Variant Information |
|---|
Single base substitutions: 51
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 11282413 | A-T | HOMO | ||
| SBS | Chr1 | 12006672 | C-T | HET | ||
| SBS | Chr1 | 12521760 | A-G | HOMO | LOC_Os01g22270 | |
| SBS | Chr1 | 30578795 | C-A | HET | ||
| SBS | Chr1 | 39351579 | G-C | HET | ||
| SBS | Chr1 | 9658842 | G-T | HET | ||
| SBS | Chr10 | 2267773 | A-G | HET | LOC_Os10g04740 | |
| SBS | Chr10 | 4262592 | G-C | HET | ||
| SBS | Chr10 | 647334 | A-G | HET | ||
| SBS | Chr10 | 985471 | C-T | HET | ||
| SBS | Chr11 | 14923124 | A-T | HOMO | ||
| SBS | Chr11 | 19539632 | T-C | HET | ||
| SBS | Chr11 | 21320933 | T-C | HET | ||
| SBS | Chr11 | 8371505 | G-T | HET | ||
| SBS | Chr12 | 11791665 | C-T | HET | ||
| SBS | Chr12 | 16700550 | T-C | HOMO | ||
| SBS | Chr12 | 17515165 | T-A | HOMO | LOC_Os12g29480 | |
| SBS | Chr12 | 21489004 | C-A | HET | ||
| SBS | Chr12 | 7896554 | G-A | HET | ||
| SBS | Chr12 | 8642449 | C-T | HET | ||
| SBS | Chr2 | 15438294 | A-C | HOMO | ||
| SBS | Chr2 | 2760422 | A-G | HET | ||
| SBS | Chr3 | 10285725 | G-A | HOMO | ||
| SBS | Chr3 | 2534645 | G-T | HOMO | LOC_Os03g05225 | |
| SBS | Chr3 | 6364936 | C-T | HET | ||
| SBS | Chr4 | 1686229 | G-C | HOMO | ||
| SBS | Chr4 | 20515751 | G-A | HET | ||
| SBS | Chr4 | 20515752 | C-G | HET | ||
| SBS | Chr4 | 20515753 | C-A | HET | ||
| SBS | Chr4 | 29763786 | A-G | HET | ||
| SBS | Chr4 | 5373145 | G-A | HET | ||
| SBS | Chr5 | 21994018 | T-G | HET | LOC_Os05g37590 | |
| SBS | Chr5 | 22285749 | T-C | HET | ||
| SBS | Chr5 | 24467434 | A-G | HET | ||
| SBS | Chr5 | 4393627 | G-A | HET | ||
| SBS | Chr6 | 13286488 | C-T | HOMO | ||
| SBS | Chr6 | 14803663 | G-A | HET | ||
| SBS | Chr6 | 24842620 | G-T | HET | ||
| SBS | Chr6 | 3770299 | G-A | HET | LOC_Os06g07770 | |
| SBS | Chr7 | 1199451 | A-G | HOMO | ||
| SBS | Chr7 | 15120548 | T-C | HET | ||
| SBS | Chr7 | 18652071 | G-C | HET | ||
| SBS | Chr7 | 25787196 | G-A | HET | ||
| SBS | Chr8 | 14790179 | C-T | HET | ||
| SBS | Chr8 | 22207010 | C-T | HET | ||
| SBS | Chr8 | 27486444 | C-T | HET | ||
| SBS | Chr8 | 3429022 | A-G | HOMO | ||
| SBS | Chr8 | 8562605 | C-A | HET | LOC_Os08g14290 | |
| SBS | Chr9 | 10962555 | C-A | HET | ||
| SBS | Chr9 | 1307374 | T-G | HET | ||
| SBS | Chr9 | 17992349 | C-T | HET |
Deletions: 30
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr2 | 2221997 | 2221997 | 1 | |
| Deletion | Chr7 | 2790144 | 2790148 | 5 | |
| Deletion | Chr4 | 2830957 | 2830957 | 1 | |
| Deletion | Chr2 | 3833301 | 3833304 | 4 | |
| Deletion | Chr9 | 4605979 | 4605980 | 2 | |
| Deletion | Chr5 | 5603717 | 5603717 | 1 | LOC_Os05g10270 |
| Deletion | Chr7 | 6527513 | 6527581 | 69 | |
| Deletion | Chr11 | 7216931 | 7217024 | 94 | |
| Deletion | Chr2 | 8742859 | 8742860 | 2 | |
| Deletion | Chr9 | 12441485 | 12441485 | 1 | |
| Deletion | Chr1 | 12553281 | 12553286 | 6 | |
| Deletion | Chr4 | 12678674 | 12678684 | 11 | |
| Deletion | Chr2 | 15625297 | 15625381 | 85 | |
| Deletion | Chr4 | 16692177 | 16692182 | 6 | |
| Deletion | Chr12 | 16712001 | 16731000 | 19000 | 3 |
| Deletion | Chr4 | 17332277 | 17332280 | 4 | |
| Deletion | Chr10 | 17579073 | 17579073 | 1 | |
| Deletion | Chr6 | 17982420 | 17982426 | 7 | |
| Deletion | Chr6 | 18053214 | 18053214 | 1 | |
| Deletion | Chr4 | 18132441 | 18132441 | 1 | |
| Deletion | Chr7 | 19225874 | 19225878 | 5 | |
| Deletion | Chr8 | 20680416 | 20680416 | 1 | |
| Deletion | Chr6 | 20730092 | 20730092 | 1 | |
| Deletion | Chr3 | 21561813 | 21561814 | 2 | |
| Deletion | Chr5 | 23592336 | 23592336 | 1 | |
| Deletion | Chr6 | 23743227 | 23743227 | 1 | LOC_Os06g39920 |
| Deletion | Chr2 | 24914955 | 24914955 | 1 | |
| Deletion | Chr1 | 25214034 | 25214034 | 1 | |
| Deletion | Chr2 | 28017054 | 28017059 | 6 | |
| Deletion | Chr2 | 32005986 | 32005986 | 1 |
Insertions: 9
No Inversion
Translocations: 5
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr4 | 1086867 | Chr1 | 617213 | |
| Translocation | Chr4 | 1087316 | Chr1 | 617202 | |
| Translocation | Chr10 | 5301589 | Chr8 | 12706298 | |
| Translocation | Chr8 | 11446160 | Chr2 | 4439077 | |
| Translocation | Chr8 | 11446161 | Chr2 | 4439402 |
