Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN4568-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN4568-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 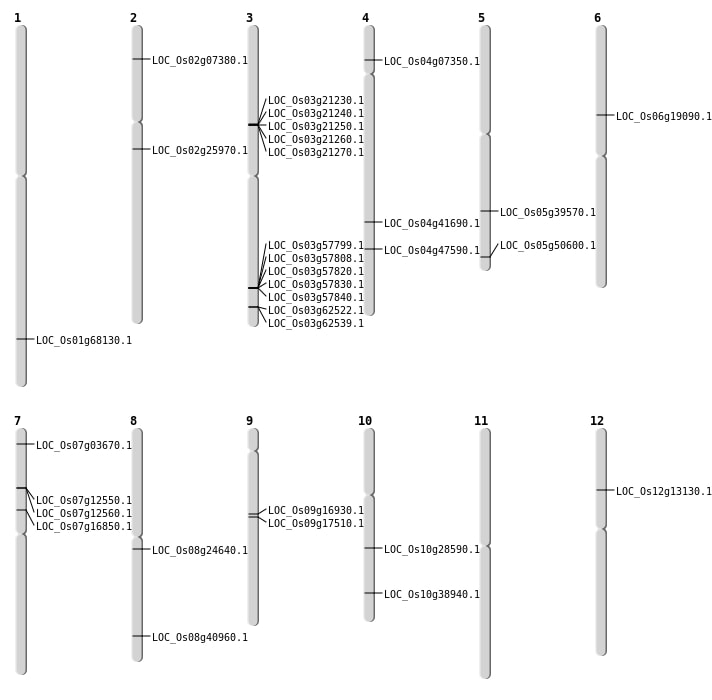
|
| Variant Information |
|---|
Single base substitutions: 43
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 11177772 | A-G | HOMO | ||
| SBS | Chr1 | 32623462 | A-T | HOMO | ||
| SBS | Chr1 | 5431889 | C-A | HET | ||
| SBS | Chr1 | 7157343 | C-T | HOMO | ||
| SBS | Chr1 | 7271365 | A-T | HET | ||
| SBS | Chr10 | 12001228 | T-A | HOMO | ||
| SBS | Chr10 | 14258731 | A-T | HOMO | ||
| SBS | Chr10 | 6452740 | G-A | HET | ||
| SBS | Chr10 | 8337962 | C-T | HET | ||
| SBS | Chr11 | 12589662 | G-T | HET | ||
| SBS | Chr12 | 14589194 | G-T | HET | ||
| SBS | Chr2 | 33486541 | T-C | HET | ||
| SBS | Chr2 | 3813163 | G-A | HET | LOC_Os02g07380 | |
| SBS | Chr3 | 11009500 | C-A | HET | ||
| SBS | Chr4 | 10083805 | C-T | HET | ||
| SBS | Chr4 | 21290955 | C-T | HET | ||
| SBS | Chr4 | 23688336 | C-T | HET | ||
| SBS | Chr4 | 29273504 | C-T | HET | ||
| SBS | Chr4 | 35434027 | C-T | HET | ||
| SBS | Chr4 | 543445 | C-T | HET | ||
| SBS | Chr4 | 8380240 | C-T | HET | ||
| SBS | Chr5 | 11794088 | G-A | HOMO | ||
| SBS | Chr5 | 13706674 | A-G | HET | ||
| SBS | Chr5 | 19123591 | T-C | HET | ||
| SBS | Chr5 | 23226998 | C-T | HET | LOC_Os05g39570 | |
| SBS | Chr5 | 29004962 | G-A | HET | LOC_Os05g50600 | |
| SBS | Chr5 | 7663366 | G-A | HET | ||
| SBS | Chr6 | 10861035 | G-T | HET | LOC_Os06g19090 | |
| SBS | Chr6 | 15024624 | G-A | HET | ||
| SBS | Chr6 | 15167187 | C-T | HET | ||
| SBS | Chr6 | 16777020 | G-T | HET | ||
| SBS | Chr6 | 17111170 | C-G | HOMO | ||
| SBS | Chr6 | 3744384 | T-A | HOMO | ||
| SBS | Chr7 | 11202939 | G-A | HOMO | ||
| SBS | Chr7 | 1900405 | A-T | HET | ||
| SBS | Chr7 | 24767661 | G-A | HET | ||
| SBS | Chr7 | 27884032 | G-T | HET | ||
| SBS | Chr7 | 7575863 | G-A | HET | ||
| SBS | Chr8 | 1406174 | A-T | HOMO | ||
| SBS | Chr8 | 17429485 | T-C | HOMO | ||
| SBS | Chr8 | 25919297 | G-A | HET | LOC_Os08g40960 | |
| SBS | Chr8 | 7511276 | A-C | HOMO | ||
| SBS | Chr9 | 19545762 | G-A | HET |
Deletions: 39
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr1 | 989424 | 989424 | 1 | |
| Deletion | Chr7 | 1488392 | 1488394 | 3 | LOC_Os07g03670 |
| Deletion | Chr12 | 2340604 | 2340607 | 4 | |
| Deletion | Chr12 | 2792546 | 2792546 | 1 | |
| Deletion | Chr7 | 3747440 | 3747441 | 2 | |
| Deletion | Chr7 | 3755566 | 3755580 | 15 | |
| Deletion | Chr4 | 3910882 | 3910887 | 6 | LOC_Os04g07350 |
| Deletion | Chr6 | 4345672 | 4345696 | 25 | |
| Deletion | Chr12 | 4515316 | 4515324 | 9 | |
| Deletion | Chr1 | 5631297 | 5631297 | 1 | |
| Deletion | Chr9 | 5770306 | 5770306 | 1 | |
| Deletion | Chr7 | 7042273 | 7042284 | 12 | |
| Deletion | Chr7 | 7055482 | 7055490 | 9 | |
| Deletion | Chr7 | 8154462 | 8154465 | 4 | |
| Deletion | Chr7 | 9906324 | 9906324 | 1 | LOC_Os07g16850 |
| Deletion | Chr1 | 10595489 | 10595491 | 3 | |
| Deletion | Chr9 | 10709948 | 10710036 | 89 | LOC_Os09g17510 |
| Deletion | Chr3 | 12111001 | 12167000 | 56000 | 5 |
| Deletion | Chr3 | 12917020 | 12917020 | 1 | |
| Deletion | Chr6 | 13237595 | 13237595 | 1 | |
| Deletion | Chr7 | 15844245 | 15844246 | 2 | |
| Deletion | Chr2 | 16523993 | 16523993 | 1 | |
| Deletion | Chr9 | 16663208 | 16663208 | 1 | |
| Deletion | Chr11 | 16995052 | 16995052 | 1 | |
| Deletion | Chr12 | 17124313 | 17124313 | 1 | |
| Deletion | Chr7 | 17905574 | 17905575 | 2 | |
| Deletion | Chr11 | 19359482 | 19359482 | 1 | |
| Deletion | Chr10 | 20745001 | 20755000 | 10000 | LOC_Os10g38940 |
| Deletion | Chr10 | 20801243 | 20801247 | 5 | |
| Deletion | Chr10 | 22543939 | 22543940 | 2 | |
| Deletion | Chr1 | 23070308 | 23070309 | 2 | |
| Deletion | Chr3 | 23970887 | 23970888 | 2 | |
| Deletion | Chr3 | 24715729 | 24715737 | 9 | |
| Deletion | Chr4 | 27944981 | 27944985 | 5 | |
| Deletion | Chr2 | 30254949 | 30254949 | 1 | LOC_Os02g49500 |
| Deletion | Chr3 | 31968717 | 31968717 | 1 | |
| Deletion | Chr3 | 32936001 | 32952000 | 16000 | 5 |
| Deletion | Chr3 | 32940836 | 32940838 | 3 | LOC_Os03g57808 |
| Deletion | Chr1 | 37069223 | 37069228 | 6 |
Insertions: 7
Inversions: 2
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr11 | 18583062 | 18583391 | |
| Inversion | Chr7 | 21168374 | 21168534 |
Translocations: 11
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr12 | 4962396 | Chr10 | 8686855 | |
| Translocation | Chr7 | 7165121 | Chr1 | 39608515 | 2 |
| Translocation | Chr7 | 7165712 | Chr1 | 39608511 | 2 |
| Translocation | Chr12 | 12069143 | Chr2 | 25830720 | |
| Translocation | Chr12 | 12069331 | Chr2 | 25830717 | |
| Translocation | Chr10 | 20745137 | Chr1 | 2891632 | LOC_Os10g38940 |
| Translocation | Chr10 | 20755276 | Chr3 | 5698770 | |
| Translocation | Chr10 | 20766518 | Chr1 | 2830179 | |
| Translocation | Chr10 | 20766645 | Chr1 | 2891660 | |
| Translocation | Chr6 | 25798698 | Chr3 | 35410876 | LOC_Os03g62539 |
| Translocation | Chr6 | 25798704 | Chr3 | 35405000 | LOC_Os03g62522 |
