Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN4571-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN4571-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 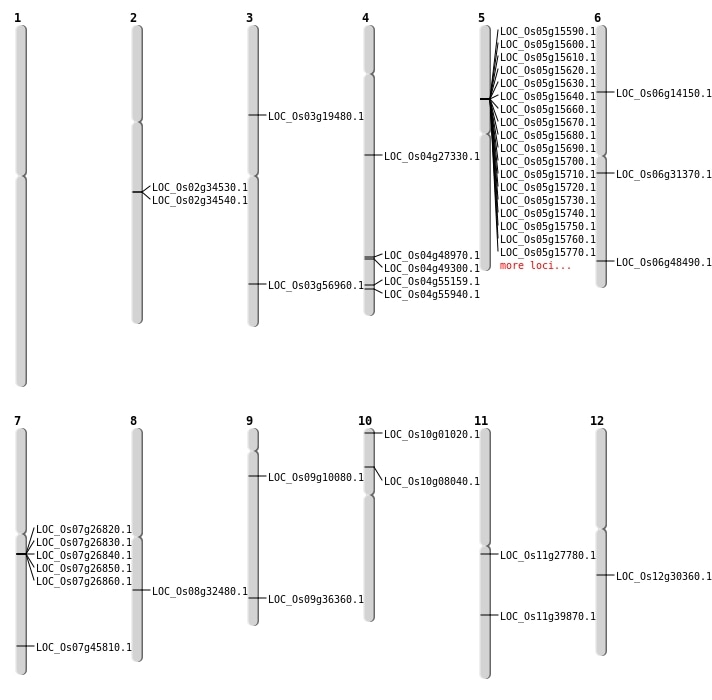
|
| Variant Information |
|---|
Single base substitutions: 50
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 13512782 | T-C | HET | ||
| SBS | Chr1 | 17833819 | G-T | HOMO | ||
| SBS | Chr1 | 19543803 | G-A | HET | ||
| SBS | Chr1 | 22821180 | C-T | HOMO | ||
| SBS | Chr1 | 42040793 | C-T | HET | ||
| SBS | Chr10 | 19396763 | T-A | HOMO | ||
| SBS | Chr10 | 2704203 | A-T | HET | ||
| SBS | Chr10 | 29161 | C-T | HET | LOC_Os10g01020 | |
| SBS | Chr10 | 7377285 | A-T | HET | ||
| SBS | Chr10 | 7770954 | T-C | HET | ||
| SBS | Chr11 | 11477660 | G-A | HET | ||
| SBS | Chr11 | 15696720 | T-G | HET | ||
| SBS | Chr11 | 23768054 | C-A | HET | LOC_Os11g39870 | |
| SBS | Chr11 | 25185337 | C-T | HET | ||
| SBS | Chr12 | 10271759 | G-T | HOMO | ||
| SBS | Chr12 | 17990219 | C-T | HET | ||
| SBS | Chr12 | 18183634 | C-A | HET | ||
| SBS | Chr12 | 24690614 | A-G | HET | ||
| SBS | Chr3 | 10953112 | T-A | HET | LOC_Os03g19480 | |
| SBS | Chr3 | 11855575 | A-T | HET | ||
| SBS | Chr3 | 18045559 | T-G | HOMO | ||
| SBS | Chr3 | 32073736 | C-A | HET | ||
| SBS | Chr3 | 32443260 | T-A | HET | LOC_Os03g56960 | |
| SBS | Chr3 | 34602041 | C-A | HET | ||
| SBS | Chr3 | 35247850 | C-T | HET | ||
| SBS | Chr4 | 11137864 | G-T | HET | ||
| SBS | Chr4 | 14570895 | C-T | HET | ||
| SBS | Chr4 | 14637932 | A-G | HET | ||
| SBS | Chr4 | 16150269 | G-A | HET | LOC_Os04g27330 | |
| SBS | Chr4 | 1738007 | G-A | HET | ||
| SBS | Chr4 | 28558098 | A-G | HET | ||
| SBS | Chr4 | 28793212 | C-T | HET | ||
| SBS | Chr4 | 33316833 | A-T | HET | LOC_Os04g55940 | |
| SBS | Chr4 | 33561659 | T-C | HET | ||
| SBS | Chr4 | 8878480 | C-T | HET | ||
| SBS | Chr5 | 10917010 | C-T | HET | ||
| SBS | Chr5 | 21350164 | G-A | HET | ||
| SBS | Chr5 | 8570097 | T-A | HET | ||
| SBS | Chr6 | 19422321 | A-C | HET | ||
| SBS | Chr6 | 19422322 | T-C | HET | ||
| SBS | Chr6 | 9865136 | C-T | HET | ||
| SBS | Chr7 | 13318507 | T-C | HET | ||
| SBS | Chr7 | 23644031 | A-C | HET | ||
| SBS | Chr8 | 20106972 | C-T | HOMO | LOC_Os08g32480 | |
| SBS | Chr8 | 20632961 | G-T | HOMO | ||
| SBS | Chr8 | 6880125 | G-A | HOMO | ||
| SBS | Chr9 | 14756658 | G-A | HOMO | ||
| SBS | Chr9 | 20693709 | A-G | HET | ||
| SBS | Chr9 | 20984043 | T-C | HOMO | LOC_Os09g36360 | |
| SBS | Chr9 | 5495450 | G-T | HOMO | LOC_Os09g10080 |
Deletions: 37
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr6 | 1316184 | 1316185 | 2 | |
| Deletion | Chr8 | 2626572 | 2626573 | 2 | |
| Deletion | Chr12 | 3465931 | 3465931 | 1 | |
| Deletion | Chr10 | 4358949 | 4358949 | 1 | LOC_Os10g08040 |
| Deletion | Chr6 | 7894842 | 7894865 | 24 | LOC_Os06g14150 |
| Deletion | Chr5 | 8837001 | 8852000 | 15000 | 6 |
| Deletion | Chr5 | 8854001 | 8891000 | 37000 | 10 |
| Deletion | Chr5 | 8894001 | 8925000 | 31000 | 7 |
| Deletion | Chr5 | 8928001 | 8994000 | 66000 | 11 |
| Deletion | Chr1 | 9188471 | 9188500 | 30 | |
| Deletion | Chr1 | 9706342 | 9706360 | 19 | |
| Deletion | Chr5 | 10059164 | 10059167 | 4 | |
| Deletion | Chr8 | 10588254 | 10588261 | 8 | |
| Deletion | Chr4 | 10818290 | 10818290 | 1 | |
| Deletion | Chr7 | 12181371 | 12181371 | 1 | |
| Deletion | Chr3 | 12925225 | 12925225 | 1 | |
| Deletion | Chr7 | 13857653 | 13857653 | 1 | |
| Deletion | Chr3 | 14861878 | 14861879 | 2 | |
| Deletion | Chr4 | 15328401 | 15328401 | 1 | |
| Deletion | Chr7 | 15508001 | 15549000 | 41000 | 5 |
| Deletion | Chr7 | 15788680 | 15788681 | 2 | |
| Deletion | Chr11 | 15990007 | 15990015 | 9 | |
| Deletion | Chr11 | 15990191 | 15990195 | 5 | LOC_Os11g27780 |
| Deletion | Chr4 | 16170604 | 16170604 | 1 | |
| Deletion | Chr12 | 18230900 | 18230900 | 1 | LOC_Os12g30360 |
| Deletion | Chr6 | 18668898 | 18668900 | 3 | |
| Deletion | Chr6 | 20123500 | 20123501 | 2 | |
| Deletion | Chr2 | 20699001 | 20707000 | 8000 | 2 |
| Deletion | Chr12 | 20762965 | 20762965 | 1 | |
| Deletion | Chr6 | 24770124 | 24770139 | 16 | |
| Deletion | Chr8 | 24857166 | 24857166 | 1 | |
| Deletion | Chr8 | 25934079 | 25934090 | 12 | |
| Deletion | Chr5 | 29371124 | 29371132 | 9 | LOC_Os05g51200 |
| Deletion | Chr5 | 29573553 | 29573568 | 16 | |
| Deletion | Chr4 | 32813218 | 32813234 | 17 | |
| Deletion | Chr4 | 35074674 | 35074676 | 3 | |
| Deletion | Chr1 | 37273308 | 37273308 | 1 |
Insertions: 20
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr1 | 11485742 | 11485743 | 2 | |
| Insertion | Chr1 | 25605462 | 25605545 | 84 | |
| Insertion | Chr10 | 22330426 | 22330426 | 1 | |
| Insertion | Chr12 | 11626172 | 11626178 | 7 | |
| Insertion | Chr12 | 16820876 | 16820876 | 1 | |
| Insertion | Chr2 | 26793248 | 26793265 | 18 | |
| Insertion | Chr2 | 31111224 | 31111265 | 42 | |
| Insertion | Chr2 | 3526193 | 3526193 | 1 | |
| Insertion | Chr3 | 33735681 | 33735681 | 1 | |
| Insertion | Chr5 | 3928278 | 3928278 | 1 | |
| Insertion | Chr6 | 1803642 | 1803642 | 1 | |
| Insertion | Chr6 | 18256280 | 18256357 | 78 | LOC_Os06g31370 |
| Insertion | Chr6 | 20213993 | 20214093 | 101 | |
| Insertion | Chr6 | 2306123 | 2306123 | 1 | |
| Insertion | Chr6 | 2586672 | 2586672 | 1 | |
| Insertion | Chr7 | 27342456 | 27342458 | 3 | LOC_Os07g45810 |
| Insertion | Chr7 | 6342962 | 6343010 | 49 | |
| Insertion | Chr8 | 18582607 | 18582681 | 75 | |
| Insertion | Chr8 | 3731944 | 3731944 | 1 | |
| Insertion | Chr9 | 16459840 | 16459840 | 1 |
Inversions: 2
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr4 | 29203460 | 29359054 | LOC_Os04g48970 |
| Inversion | Chr4 | 29359051 | 29426273 | LOC_Os04g49300 |
No Translocation
