Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN4573-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN4573-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 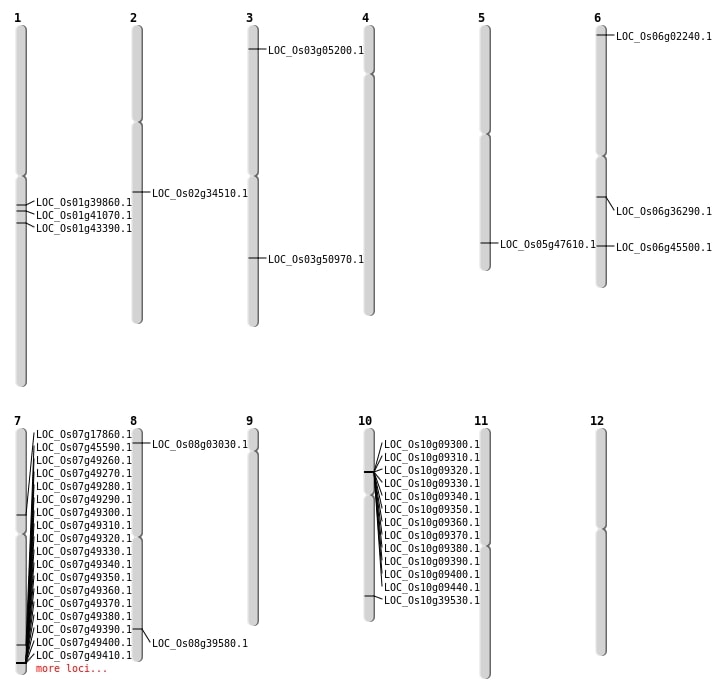
|
| Variant Information |
|---|
Single base substitutions: 49
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 16398004 | G-A | HET | ||
| SBS | Chr1 | 22490059 | G-C | HET | LOC_Os01g39860 | |
| SBS | Chr1 | 23260839 | A-C | HET | LOC_Os01g41070 | |
| SBS | Chr1 | 25247238 | A-T | HET | ||
| SBS | Chr1 | 37876195 | C-T | HET | ||
| SBS | Chr1 | 7688743 | C-T | HET | ||
| SBS | Chr10 | 14791592 | G-C | HET | ||
| SBS | Chr10 | 1583100 | T-A | HET | ||
| SBS | Chr10 | 21119134 | G-C | HET | LOC_Os10g39530 | |
| SBS | Chr11 | 17533093 | G-T | HET | ||
| SBS | Chr11 | 8591059 | G-A | HET | ||
| SBS | Chr11 | 8729359 | A-G | HET | ||
| SBS | Chr12 | 13807976 | T-C | HET | ||
| SBS | Chr12 | 18306537 | T-C | HOMO | ||
| SBS | Chr12 | 26566763 | T-C | HET | ||
| SBS | Chr2 | 25598299 | G-A | HOMO | ||
| SBS | Chr2 | 34415509 | C-T | HET | ||
| SBS | Chr2 | 35687137 | C-T | HET | ||
| SBS | Chr2 | 6259069 | T-C | HOMO | ||
| SBS | Chr2 | 6840779 | C-A | HOMO | ||
| SBS | Chr3 | 17272599 | G-T | HET | ||
| SBS | Chr3 | 257470 | G-A | HET | ||
| SBS | Chr3 | 4408749 | T-C | HET | ||
| SBS | Chr4 | 10384185 | G-A | HOMO | ||
| SBS | Chr4 | 13163151 | T-C | HET | ||
| SBS | Chr4 | 18231697 | G-A | HET | ||
| SBS | Chr4 | 25948529 | T-A | HET | ||
| SBS | Chr4 | 8379179 | A-T | HOMO | ||
| SBS | Chr5 | 12427621 | G-T | HET | ||
| SBS | Chr5 | 19228285 | T-A | HET | ||
| SBS | Chr5 | 20440863 | G-T | HET | ||
| SBS | Chr5 | 20440864 | C-T | HET | ||
| SBS | Chr5 | 24320040 | T-A | HET | ||
| SBS | Chr5 | 25120877 | C-T | HET | ||
| SBS | Chr5 | 27280118 | G-A | HET | LOC_Os05g47610 | |
| SBS | Chr6 | 16338826 | C-T | HET | ||
| SBS | Chr6 | 17070541 | C-T | HOMO | ||
| SBS | Chr6 | 19499263 | C-T | HET | ||
| SBS | Chr6 | 24905991 | C-G | HET | ||
| SBS | Chr7 | 13861386 | C-T | HET | ||
| SBS | Chr7 | 15229266 | A-G | HOMO | ||
| SBS | Chr7 | 27209611 | G-A | HET | LOC_Os07g45590 | |
| SBS | Chr8 | 22224104 | G-T | HET | ||
| SBS | Chr8 | 25044040 | G-A | HET | LOC_Os08g39580 | |
| SBS | Chr8 | 25265315 | G-A | HET | ||
| SBS | Chr8 | 26040590 | G-C | HET | ||
| SBS | Chr8 | 27243242 | C-T | HOMO | ||
| SBS | Chr8 | 2780571 | A-G | HET | ||
| SBS | Chr8 | 3297725 | A-C | HET |
Deletions: 30
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr8 | 355579 | 355585 | 7 | |
| Deletion | Chr10 | 461059 | 461060 | 2 | |
| Deletion | Chr6 | 1051742 | 1051744 | 3 | |
| Deletion | Chr3 | 2524471 | 2524471 | 1 | LOC_Os03g05200 |
| Deletion | Chr3 | 2630261 | 2630269 | 9 | |
| Deletion | Chr10 | 5028001 | 5038000 | 10000 | 2 |
| Deletion | Chr10 | 5040001 | 5072000 | 32000 | 9 |
| Deletion | Chr10 | 5093091 | 5093096 | 6 | LOC_Os10g09440 |
| Deletion | Chr10 | 5104929 | 5104929 | 1 | |
| Deletion | Chr7 | 7456610 | 7456628 | 19 | |
| Deletion | Chr10 | 8098027 | 8098055 | 29 | |
| Deletion | Chr1 | 9486621 | 9486625 | 5 | |
| Deletion | Chr1 | 11485189 | 11485194 | 6 | |
| Deletion | Chr7 | 11606100 | 11606104 | 5 | |
| Deletion | Chr12 | 12406026 | 12406028 | 3 | |
| Deletion | Chr12 | 12434519 | 12434519 | 1 | |
| Deletion | Chr4 | 12627976 | 12627976 | 1 | |
| Deletion | Chr12 | 19262065 | 19262067 | 3 | |
| Deletion | Chr1 | 20028734 | 20028736 | 3 | |
| Deletion | Chr5 | 20636473 | 20636475 | 3 | |
| Deletion | Chr8 | 22073984 | 22073984 | 1 | |
| Deletion | Chr3 | 23215136 | 23215158 | 23 | |
| Deletion | Chr5 | 24020614 | 24020625 | 12 | |
| Deletion | Chr5 | 24531041 | 24531042 | 2 | |
| Deletion | Chr7 | 24688080 | 24688080 | 1 | |
| Deletion | Chr5 | 25605537 | 25605537 | 1 | |
| Deletion | Chr4 | 25653126 | 25653138 | 13 | |
| Deletion | Chr12 | 25951140 | 25951140 | 1 | |
| Deletion | Chr8 | 28271028 | 28271031 | 4 | |
| Deletion | Chr7 | 29493001 | 29671000 | 178000 | 27 |
Insertions: 13
Inversions: 5
Translocations: 7
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr6 | 702136 | Chr2 | 22762716 | LOC_Os06g02240 |
| Translocation | Chr6 | 702144 | Chr2 | 22763473 | LOC_Os06g02240 |
| Translocation | Chr8 | 1127270 | Chr1 | 25265670 | |
| Translocation | Chr8 | 1356823 | Chr1 | 25266306 | LOC_Os08g03030 |
| Translocation | Chr12 | 7388810 | Chr10 | 3079686 | |
| Translocation | Chr4 | 22487068 | Chr3 | 29119853 | LOC_Os03g50970 |
| Translocation | Chr4 | 22487071 | Chr3 | 29119958 |
