Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN4575-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN4575-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 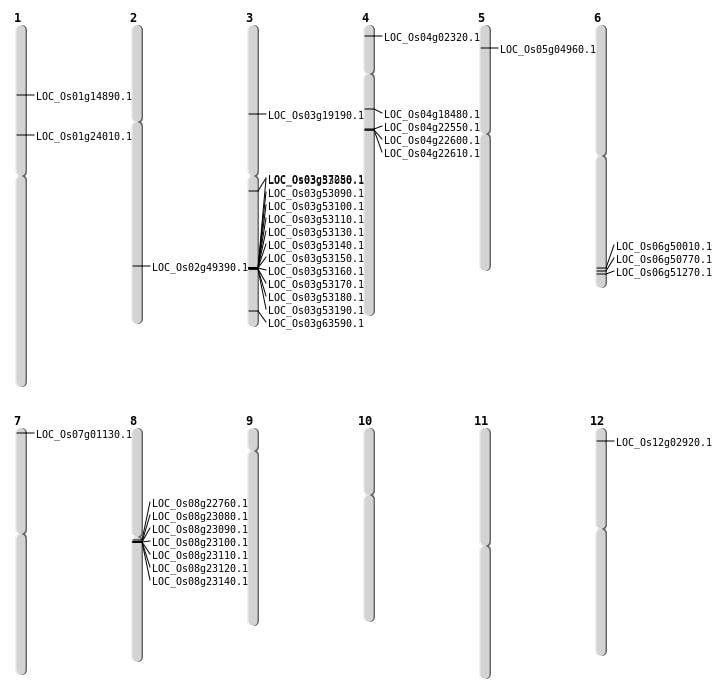
|
| Variant Information |
|---|
Single base substitutions: 65
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 13524638 | G-A | HET | LOC_Os01g24010 | |
| SBS | Chr1 | 20578144 | T-C | HET | ||
| SBS | Chr1 | 22665779 | C-A | HET | ||
| SBS | Chr1 | 23989238 | C-A | HET | ||
| SBS | Chr1 | 25800607 | G-T | HET | ||
| SBS | Chr1 | 26822908 | C-T | HET | ||
| SBS | Chr1 | 37443740 | C-T | HET | ||
| SBS | Chr1 | 41678286 | A-G | HET | ||
| SBS | Chr10 | 19648246 | G-A | HOMO | ||
| SBS | Chr10 | 274733 | T-C | HOMO | ||
| SBS | Chr10 | 5126631 | C-T | HOMO | ||
| SBS | Chr11 | 16617092 | C-G | HOMO | ||
| SBS | Chr11 | 6020147 | C-G | HOMO | ||
| SBS | Chr12 | 12209053 | C-T | HET | ||
| SBS | Chr12 | 12709273 | A-G | HET | ||
| SBS | Chr12 | 16283365 | G-A | HET | ||
| SBS | Chr12 | 19929441 | C-T | HOMO | ||
| SBS | Chr12 | 5608010 | C-A | HET | ||
| SBS | Chr2 | 21092145 | C-G | HET | ||
| SBS | Chr2 | 24008839 | T-A | HET | ||
| SBS | Chr2 | 30176294 | C-T | HET | LOC_Os02g49390 | |
| SBS | Chr2 | 30617011 | G-A | HET | ||
| SBS | Chr2 | 31999547 | C-G | HET | ||
| SBS | Chr2 | 31999548 | G-T | HET | ||
| SBS | Chr3 | 10755660 | G-A | HET | LOC_Os03g19190 | |
| SBS | Chr3 | 20638230 | A-G | HET | ||
| SBS | Chr3 | 20647441 | T-A | HET | LOC_Os03g37250 | |
| SBS | Chr3 | 24085412 | C-T | HET | ||
| SBS | Chr3 | 5567347 | G-C | HET | ||
| SBS | Chr4 | 10141948 | C-T | HET | ||
| SBS | Chr4 | 12769271 | G-A | HET | LOC_Os04g22550 | |
| SBS | Chr4 | 12806707 | A-G | HET | ||
| SBS | Chr4 | 12809122 | G-C | HET | LOC_Os04g22610 | |
| SBS | Chr4 | 13888214 | G-A | HET | ||
| SBS | Chr4 | 16830446 | C-A | HET | ||
| SBS | Chr4 | 26758915 | C-T | HET | ||
| SBS | Chr4 | 3270869 | T-C | HET | ||
| SBS | Chr4 | 8648090 | G-A | HET | ||
| SBS | Chr5 | 15136080 | G-T | HET | ||
| SBS | Chr5 | 17745704 | T-A | HET | ||
| SBS | Chr5 | 17745705 | C-T | HET | ||
| SBS | Chr5 | 20860796 | C-T | HOMO | ||
| SBS | Chr5 | 23114197 | T-C | HOMO | ||
| SBS | Chr5 | 23403881 | G-T | HOMO | ||
| SBS | Chr5 | 2400855 | C-A | HET | LOC_Os05g04960 | |
| SBS | Chr6 | 29678325 | G-A | HET | ||
| SBS | Chr6 | 30285103 | G-T | HET | LOC_Os06g50010 | |
| SBS | Chr6 | 30285104 | C-T | HET | LOC_Os06g50010 | |
| SBS | Chr6 | 31035050 | G-A | HET | LOC_Os06g51270 | |
| SBS | Chr7 | 10660161 | T-A | HET | ||
| SBS | Chr7 | 5721039 | G-A | HET | ||
| SBS | Chr7 | 74422 | G-A | HET | LOC_Os07g01130 | |
| SBS | Chr7 | 8827876 | A-T | HOMO | ||
| SBS | Chr8 | 14101762 | T-G | HET | ||
| SBS | Chr8 | 14101770 | C-T | HET | ||
| SBS | Chr8 | 22384446 | T-C | HET | ||
| SBS | Chr9 | 13374951 | A-T | HET | ||
| SBS | Chr9 | 13628616 | A-T | HET | ||
| SBS | Chr9 | 17994526 | G-T | HET | ||
| SBS | Chr9 | 5487120 | T-A | HOMO | ||
| SBS | Chr9 | 5487121 | C-A | HOMO | ||
| SBS | Chr9 | 5625006 | A-G | HOMO | ||
| SBS | Chr9 | 5681522 | T-C | HET | ||
| SBS | Chr9 | 6583001 | T-C | HET | ||
| SBS | Chr9 | 7437086 | A-G | HOMO |
Deletions: 23
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr11 | 1314498 | 1314505 | 8 | |
| Deletion | Chr6 | 4312301 | 4312316 | 16 | |
| Deletion | Chr8 | 8183095 | 8183095 | 1 | |
| Deletion | Chr1 | 8344549 | 8344554 | 6 | LOC_Os01g14890 |
| Deletion | Chr9 | 12458645 | 12458650 | 6 | |
| Deletion | Chr4 | 12638968 | 12638968 | 1 | |
| Deletion | Chr4 | 12794001 | 12806000 | 12000 | LOC_Os04g22600 |
| Deletion | Chr8 | 13903001 | 13945000 | 42000 | 7 |
| Deletion | Chr8 | 14091278 | 14091293 | 16 | |
| Deletion | Chr8 | 14523868 | 14523869 | 2 | |
| Deletion | Chr2 | 14886599 | 14886599 | 1 | |
| Deletion | Chr10 | 16002341 | 16002341 | 1 | |
| Deletion | Chr9 | 18384915 | 18384917 | 3 | |
| Deletion | Chr12 | 19984094 | 19984099 | 6 | |
| Deletion | Chr8 | 22123292 | 22123310 | 19 | |
| Deletion | Chr12 | 22447451 | 22447455 | 5 | |
| Deletion | Chr5 | 22478157 | 22478175 | 19 | |
| Deletion | Chr4 | 23130590 | 23130590 | 1 | |
| Deletion | Chr1 | 23727954 | 23727954 | 1 | |
| Deletion | Chr4 | 24802459 | 24802473 | 15 | |
| Deletion | Chr6 | 27034649 | 27034649 | 1 | |
| Deletion | Chr11 | 27968403 | 27968411 | 9 | |
| Deletion | Chr3 | 30444001 | 30514000 | 70000 | 11 |
Insertions: 13
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr1 | 11799491 | 11799585 | 95 | |
| Insertion | Chr11 | 15152328 | 15152328 | 1 | |
| Insertion | Chr11 | 28651415 | 28651484 | 70 | |
| Insertion | Chr12 | 1081945 | 1081947 | 3 | LOC_Os12g02920 |
| Insertion | Chr12 | 22116317 | 22116322 | 6 | |
| Insertion | Chr2 | 1257549 | 1257553 | 5 | |
| Insertion | Chr2 | 17825854 | 17825854 | 1 | |
| Insertion | Chr2 | 32563716 | 32563716 | 1 | |
| Insertion | Chr2 | 9197948 | 9197948 | 1 | |
| Insertion | Chr4 | 13773155 | 13773158 | 4 | |
| Insertion | Chr5 | 26230330 | 26230330 | 1 | |
| Insertion | Chr8 | 3252362 | 3252363 | 2 | |
| Insertion | Chr9 | 22344583 | 22344606 | 24 |
Inversions: 4
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr4 | 745220 | 814095 | LOC_Os04g02320 |
| Inversion | Chr4 | 16189455 | 16189653 | |
| Inversion | Chr6 | 22660296 | 23213894 | |
| Inversion | Chr6 | 22660307 | 23213898 |
Translocations: 5
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr11 | 3393128 | Chr9 | 13679315 | |
| Translocation | Chr11 | 3393600 | Chr9 | 16985189 | |
| Translocation | Chr5 | 6775394 | Chr4 | 745200 | |
| Translocation | Chr8 | 13684523 | Chr4 | 10213071 | 2 |
| Translocation | Chr12 | 17125522 | Chr9 | 5613220 |
