Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN4577-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN4577-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 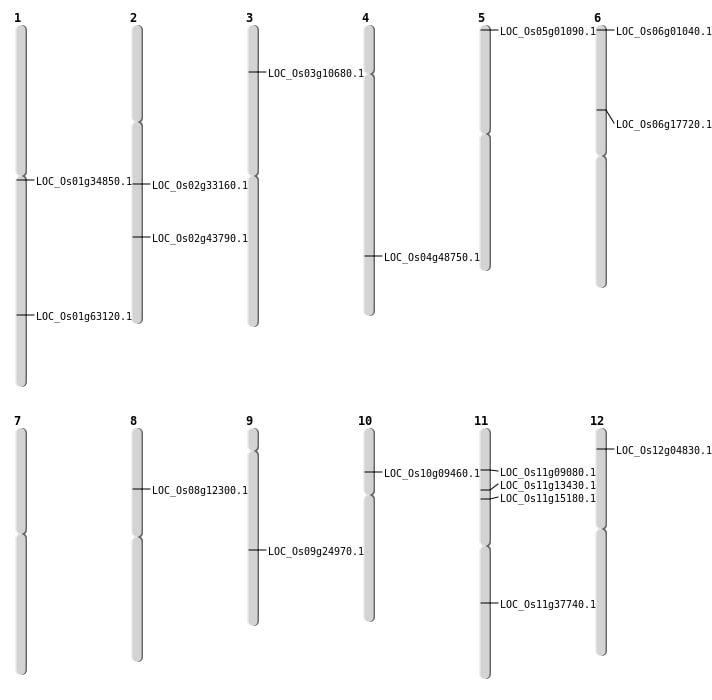
|
| Variant Information |
|---|
Single base substitutions: 39
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 30371060 | G-A | HET | ||
| SBS | Chr1 | 40628429 | G-A | HOMO | ||
| SBS | Chr1 | 40794924 | A-T | HOMO | ||
| SBS | Chr1 | 5231829 | C-T | HET | ||
| SBS | Chr1 | 685940 | G-C | HET | ||
| SBS | Chr1 | 7652125 | C-T | HOMO | ||
| SBS | Chr10 | 14831817 | G-A | HOMO | ||
| SBS | Chr10 | 16647766 | C-T | HET | ||
| SBS | Chr10 | 811150 | C-T | HOMO | ||
| SBS | Chr10 | 9563716 | G-A | HET | ||
| SBS | Chr11 | 22315480 | C-T | HET | LOC_Os11g37740 | |
| SBS | Chr11 | 23662139 | C-T | HET | ||
| SBS | Chr12 | 13395882 | C-T | HET | ||
| SBS | Chr12 | 2072197 | G-A | HET | LOC_Os12g04830 | |
| SBS | Chr12 | 25880939 | C-T | HET | ||
| SBS | Chr2 | 10905833 | G-C | HOMO | ||
| SBS | Chr2 | 26423142 | G-A | HOMO | LOC_Os02g43790 | |
| SBS | Chr2 | 34745276 | G-A | HOMO | ||
| SBS | Chr2 | 35520546 | G-A | HET | ||
| SBS | Chr3 | 11063838 | C-T | HET | ||
| SBS | Chr3 | 12345744 | A-C | HOMO | ||
| SBS | Chr3 | 19165014 | A-T | HOMO | ||
| SBS | Chr3 | 6405043 | C-T | HET | ||
| SBS | Chr4 | 23789074 | A-T | HET | ||
| SBS | Chr4 | 3193082 | C-G | HOMO | ||
| SBS | Chr6 | 14075131 | A-G | HOMO | ||
| SBS | Chr6 | 20165192 | G-A | HOMO | ||
| SBS | Chr7 | 23998368 | G-A | HET | ||
| SBS | Chr7 | 7844806 | C-T | HOMO | ||
| SBS | Chr7 | 9292859 | C-A | HET | ||
| SBS | Chr8 | 22865101 | C-T | HET | ||
| SBS | Chr8 | 24816658 | G-T | HET | ||
| SBS | Chr8 | 24816659 | A-T | HET | ||
| SBS | Chr8 | 25884364 | C-T | HET | ||
| SBS | Chr8 | 3888860 | G-A | HET | ||
| SBS | Chr8 | 679829 | A-T | HET | ||
| SBS | Chr8 | 7248779 | G-A | HET | LOC_Os08g12300 | |
| SBS | Chr9 | 14429432 | G-A | HOMO | ||
| SBS | Chr9 | 14918145 | C-T | HOMO | LOC_Os09g24970 |
Deletions: 34
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr8 | 316691 | 316691 | 1 | |
| Deletion | Chr10 | 1255092 | 1255092 | 1 | |
| Deletion | Chr8 | 2488402 | 2488460 | 59 | |
| Deletion | Chr9 | 4752489 | 4752504 | 16 | |
| Deletion | Chr5 | 4963797 | 4963806 | 10 | |
| Deletion | Chr10 | 5101993 | 5101998 | 6 | LOC_Os10g09460 |
| Deletion | Chr3 | 5126245 | 5126245 | 1 | |
| Deletion | Chr2 | 5496717 | 5496740 | 24 | |
| Deletion | Chr4 | 5671722 | 5671722 | 1 | |
| Deletion | Chr3 | 6645563 | 6645563 | 1 | |
| Deletion | Chr11 | 7265513 | 7265513 | 1 | |
| Deletion | Chr11 | 7359218 | 7359224 | 7 | LOC_Os11g13430 |
| Deletion | Chr6 | 7748723 | 7748731 | 9 | |
| Deletion | Chr6 | 10286510 | 10286528 | 19 | LOC_Os06g17720 |
| Deletion | Chr9 | 12694665 | 12694678 | 14 | |
| Deletion | Chr8 | 13644875 | 13644875 | 1 | |
| Deletion | Chr1 | 13815679 | 13815680 | 2 | |
| Deletion | Chr8 | 14090616 | 14090688 | 73 | |
| Deletion | Chr9 | 14918160 | 14918163 | 4 | LOC_Os09g24970 |
| Deletion | Chr3 | 15549832 | 15549832 | 1 | |
| Deletion | Chr12 | 16030093 | 16030093 | 1 | |
| Deletion | Chr7 | 16870530 | 16870533 | 4 | |
| Deletion | Chr6 | 17293178 | 17293195 | 18 | |
| Deletion | Chr2 | 18769626 | 18769626 | 1 | |
| Deletion | Chr3 | 19475341 | 19475341 | 1 | |
| Deletion | Chr12 | 24418299 | 24418302 | 4 | |
| Deletion | Chr6 | 26081981 | 26081994 | 14 | |
| Deletion | Chr1 | 26553398 | 26553399 | 2 | |
| Deletion | Chr4 | 29553725 | 29553725 | 1 | |
| Deletion | Chr4 | 29708752 | 29708752 | 1 | |
| Deletion | Chr1 | 31247855 | 31247856 | 2 | |
| Deletion | Chr1 | 36576610 | 36576612 | 3 | LOC_Os01g63120 |
| Deletion | Chr1 | 37215854 | 37215865 | 12 | |
| Deletion | Chr1 | 37697764 | 37697764 | 1 |
Insertions: 17
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr1 | 1099682 | 1099687 | 6 | |
| Insertion | Chr1 | 42536480 | 42536481 | 2 | |
| Insertion | Chr1 | 7913167 | 7913167 | 1 | |
| Insertion | Chr10 | 15079982 | 15079985 | 4 | |
| Insertion | Chr10 | 2142781 | 2142781 | 1 | |
| Insertion | Chr11 | 4338806 | 4338816 | 11 | |
| Insertion | Chr11 | 7140641 | 7140642 | 2 | |
| Insertion | Chr11 | 8563818 | 8563828 | 11 | LOC_Os11g15180 |
| Insertion | Chr2 | 19722099 | 19722099 | 1 | LOC_Os02g33160 |
| Insertion | Chr2 | 33684069 | 33684070 | 2 | |
| Insertion | Chr3 | 10340807 | 10340907 | 101 | |
| Insertion | Chr3 | 14672323 | 14672404 | 82 | |
| Insertion | Chr4 | 5456370 | 5456370 | 1 | |
| Insertion | Chr5 | 25978992 | 25978992 | 1 | |
| Insertion | Chr6 | 14261124 | 14261128 | 5 | |
| Insertion | Chr7 | 21915475 | 21915475 | 1 | |
| Insertion | Chr7 | 21969070 | 21969071 | 2 |
Inversions: 3
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr12 | 25208823 | 25586502 | |
| Inversion | Chr12 | 25208831 | 25586495 | |
| Inversion | Chr4 | 29073784 | 29074189 | LOC_Os04g48750 |
Translocations: 1
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr11 | 4822263 | Chr5 | 29699490 | LOC_Os11g09080 |
