Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN4591-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN4591-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 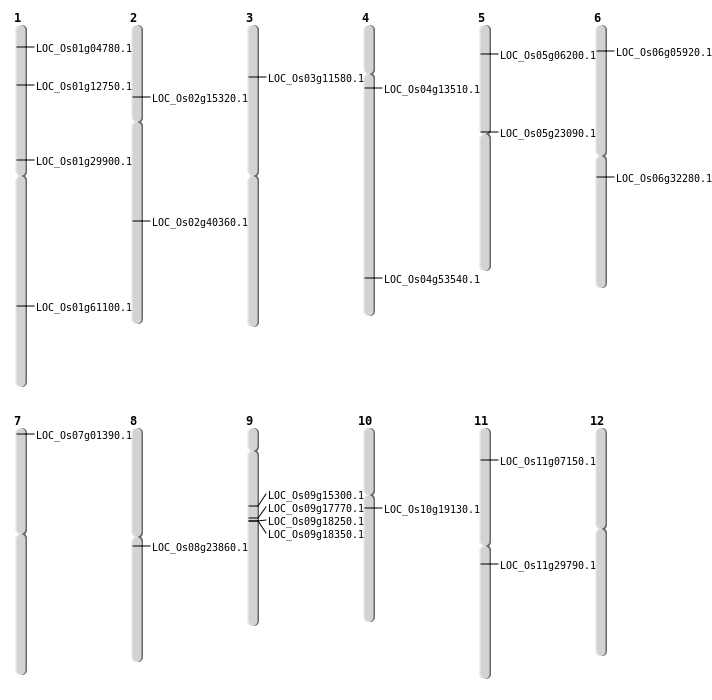
|
| Variant Information |
|---|
Single base substitutions: 44
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 2084040 | C-T | HET | ||
| SBS | Chr1 | 38039738 | A-G | HET | ||
| SBS | Chr1 | 42102183 | C-T | HET | ||
| SBS | Chr10 | 19973271 | G-A | HET | ||
| SBS | Chr10 | 3788314 | A-G | HOMO | ||
| SBS | Chr10 | 46315 | G-T | HET | ||
| SBS | Chr10 | 7350933 | C-T | HOMO | ||
| SBS | Chr10 | 7972683 | T-A | HET | ||
| SBS | Chr11 | 17292262 | T-A | HET | LOC_Os11g29790 | |
| SBS | Chr11 | 27230675 | G-A | HET | ||
| SBS | Chr11 | 3563877 | G-A | HET | LOC_Os11g07150 | |
| SBS | Chr12 | 15114081 | T-C | HET | ||
| SBS | Chr12 | 3104993 | G-A | HET | ||
| SBS | Chr2 | 19524884 | A-G | HET | ||
| SBS | Chr2 | 3702563 | T-A | HET | ||
| SBS | Chr3 | 26644315 | G-A | HET | ||
| SBS | Chr3 | 29165918 | C-T | HET | ||
| SBS | Chr3 | 30292436 | G-A | HET | ||
| SBS | Chr4 | 13079747 | C-T | HET | ||
| SBS | Chr4 | 26111567 | G-A | HET | ||
| SBS | Chr4 | 31584485 | G-A | HET | ||
| SBS | Chr4 | 31901385 | C-T | HET | LOC_Os04g53540 | |
| SBS | Chr4 | 32485714 | G-T | HOMO | ||
| SBS | Chr4 | 33324882 | T-A | HET | ||
| SBS | Chr4 | 34223552 | T-C | HET | ||
| SBS | Chr5 | 18269783 | G-A | HET | ||
| SBS | Chr5 | 28510500 | G-T | HOMO | ||
| SBS | Chr5 | 3122797 | G-A | HET | LOC_Os05g06200 | |
| SBS | Chr6 | 15097031 | A-G | HET | ||
| SBS | Chr6 | 25062507 | C-T | HET | ||
| SBS | Chr6 | 2711453 | G-A | HET | LOC_Os06g05920 | |
| SBS | Chr6 | 7082611 | T-C | HET | ||
| SBS | Chr7 | 12846118 | A-G | HOMO | ||
| SBS | Chr7 | 25464062 | G-A | HET | ||
| SBS | Chr8 | 16860977 | T-C | HET | ||
| SBS | Chr8 | 3860091 | C-T | HET | ||
| SBS | Chr8 | 415886 | C-G | HET | ||
| SBS | Chr8 | 5136711 | A-G | HET | ||
| SBS | Chr9 | 11203953 | G-A | HOMO | LOC_Os09g18250 | |
| SBS | Chr9 | 15530549 | T-C | HOMO | ||
| SBS | Chr9 | 22254353 | C-G | HOMO | ||
| SBS | Chr9 | 8374613 | C-T | HOMO | ||
| SBS | Chr9 | 9319068 | T-C | HOMO | ||
| SBS | Chr9 | 941497 | C-G | HOMO |
Deletions: 33
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr7 | 243070 | 243071 | 2 | LOC_Os07g01390 |
| Deletion | Chr7 | 1260723 | 1261133 | 411 | |
| Deletion | Chr1 | 2187967 | 2187967 | 1 | LOC_Os01g04780 |
| Deletion | Chr2 | 2225293 | 2225303 | 11 | |
| Deletion | Chr4 | 3365716 | 3365716 | 1 | |
| Deletion | Chr6 | 3815426 | 3815426 | 1 | |
| Deletion | Chr5 | 3906987 | 3906988 | 2 | |
| Deletion | Chr1 | 6988970 | 6988970 | 1 | |
| Deletion | Chr4 | 7532229 | 7532230 | 2 | LOC_Os04g13510 |
| Deletion | Chr1 | 8316543 | 8316548 | 6 | |
| Deletion | Chr6 | 8536018 | 8536018 | 1 | |
| Deletion | Chr2 | 8567501 | 8567501 | 1 | LOC_Os02g15320 |
| Deletion | Chr10 | 8584110 | 8584376 | 267 | |
| Deletion | Chr9 | 9334640 | 9334642 | 3 | LOC_Os09g15300 |
| Deletion | Chr12 | 9612865 | 9612875 | 11 | |
| Deletion | Chr12 | 9978561 | 9978704 | 144 | |
| Deletion | Chr9 | 10879105 | 10879108 | 4 | |
| Deletion | Chr9 | 11649525 | 11649567 | 43 | |
| Deletion | Chr11 | 12097975 | 12097977 | 3 | |
| Deletion | Chr11 | 13237231 | 13237256 | 26 | |
| Deletion | Chr2 | 13705399 | 13705553 | 155 | |
| Deletion | Chr1 | 14672426 | 14672426 | 1 | |
| Deletion | Chr10 | 15249197 | 15249275 | 79 | |
| Deletion | Chr1 | 15848786 | 15848786 | 1 | |
| Deletion | Chr6 | 15988354 | 15988355 | 2 | |
| Deletion | Chr3 | 16033765 | 16033766 | 2 | |
| Deletion | Chr12 | 20570451 | 20570460 | 10 | |
| Deletion | Chr2 | 21836679 | 21836679 | 1 | |
| Deletion | Chr8 | 22474645 | 22474678 | 34 | |
| Deletion | Chr2 | 22797259 | 22797281 | 23 | |
| Deletion | Chr5 | 24127542 | 24127543 | 2 | |
| Deletion | Chr5 | 28223010 | 28223010 | 1 | |
| Deletion | Chr1 | 42427852 | 42427852 | 1 |
Insertions: 15
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr1 | 12479643 | 12479644 | 2 | |
| Insertion | Chr1 | 16763248 | 16763250 | 3 | LOC_Os01g29900 |
| Insertion | Chr10 | 8752996 | 8752996 | 1 | |
| Insertion | Chr11 | 2958143 | 2958143 | 1 | |
| Insertion | Chr12 | 6821750 | 6821788 | 39 | |
| Insertion | Chr2 | 9360439 | 9360443 | 5 | |
| Insertion | Chr4 | 5581395 | 5581434 | 40 | |
| Insertion | Chr5 | 13166757 | 13166757 | 1 | LOC_Os05g23090 |
| Insertion | Chr6 | 11662289 | 11662329 | 41 | |
| Insertion | Chr7 | 19872782 | 19872782 | 1 | |
| Insertion | Chr8 | 16171416 | 16171416 | 1 | |
| Insertion | Chr8 | 18634007 | 18634007 | 1 | |
| Insertion | Chr8 | 2083191 | 2083254 | 64 | |
| Insertion | Chr8 | 6527256 | 6527258 | 3 | |
| Insertion | Chr9 | 5495803 | 5495804 | 2 |
Inversions: 3
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr2 | 7336601 | 7336752 | |
| Inversion | Chr9 | 10877946 | 11255109 | 2 |
| Inversion | Chr9 | 10877954 | 11255114 | 2 |
Translocations: 2
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr9 | 13746600 | Chr6 | 19186290 | |
| Translocation | Chr4 | 29626136 | Chr2 | 35571986 |
