Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN4595-2-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN4595-2-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 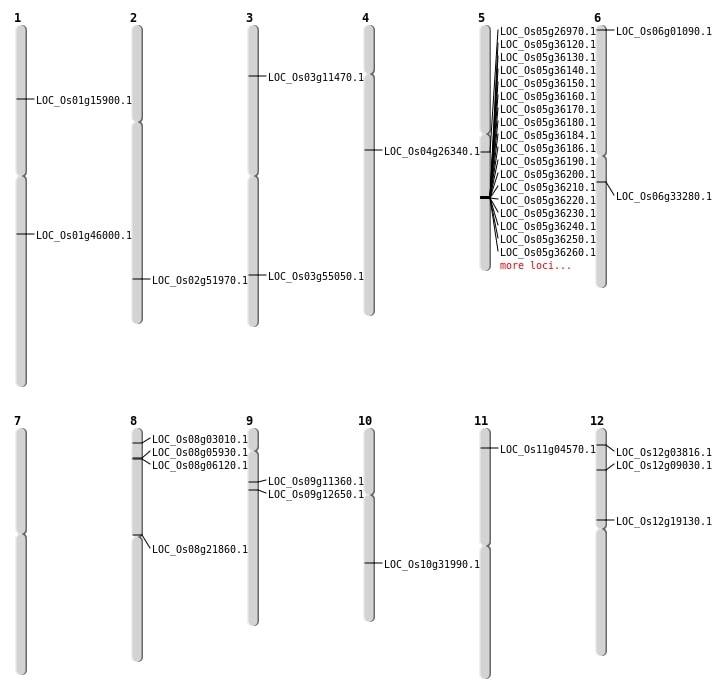
|
| Variant Information |
|---|
Single base substitutions: 43
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 13235600 | T-C | HET | ||
| SBS | Chr1 | 23659329 | G-A | HET | ||
| SBS | Chr1 | 25906165 | G-A | HET | ||
| SBS | Chr1 | 26146390 | G-A | HET | LOC_Os01g46000 | |
| SBS | Chr1 | 5638058 | C-T | HET | ||
| SBS | Chr1 | 9886578 | C-T | HET | ||
| SBS | Chr10 | 15210636 | T-A | HOMO | ||
| SBS | Chr10 | 5427356 | C-T | HET | ||
| SBS | Chr10 | 864926 | A-C | HET | ||
| SBS | Chr12 | 16184264 | G-A | HET | ||
| SBS | Chr12 | 21595981 | A-G | HET | ||
| SBS | Chr12 | 24115026 | A-T | HET | ||
| SBS | Chr12 | 24662667 | C-T | HET | ||
| SBS | Chr12 | 2804319 | C-T | HOMO | ||
| SBS | Chr12 | 3786427 | C-T | HOMO | ||
| SBS | Chr12 | 3786428 | T-C | HOMO | ||
| SBS | Chr2 | 32774894 | G-A | HOMO | ||
| SBS | Chr2 | 33891846 | A-G | HET | ||
| SBS | Chr2 | 5880555 | G-T | HET | ||
| SBS | Chr2 | 5880556 | A-T | HET | ||
| SBS | Chr3 | 17133851 | T-C | HET | ||
| SBS | Chr3 | 19808984 | G-A | HOMO | ||
| SBS | Chr3 | 31302778 | T-C | HET | LOC_Os03g55050 | |
| SBS | Chr3 | 8774120 | A-G | HET | ||
| SBS | Chr3 | 9367822 | G-A | HET | ||
| SBS | Chr4 | 12439917 | G-A | HET | ||
| SBS | Chr4 | 20226224 | A-G | HET | ||
| SBS | Chr4 | 28347007 | A-T | HET | ||
| SBS | Chr4 | 8640138 | T-A | HET | ||
| SBS | Chr5 | 12787228 | T-C | HET | ||
| SBS | Chr5 | 21404122 | C-T | HOMO | ||
| SBS | Chr5 | 21404125 | C-T | HOMO | ||
| SBS | Chr5 | 21404126 | C-T | HOMO | ||
| SBS | Chr5 | 28670551 | G-A | HET | ||
| SBS | Chr5 | 29075501 | C-G | HET | ||
| SBS | Chr6 | 1619971 | C-T | HOMO | ||
| SBS | Chr6 | 5944864 | A-G | HET | ||
| SBS | Chr6 | 99272 | T-C | HET | LOC_Os06g01090 | |
| SBS | Chr8 | 13087350 | T-A | HET | LOC_Os08g21860 | |
| SBS | Chr8 | 24290051 | T-C | HET | ||
| SBS | Chr8 | 3305371 | A-G | HET | ||
| SBS | Chr9 | 10234698 | C-T | HET | ||
| SBS | Chr9 | 6310652 | C-T | HET | LOC_Os09g11360 |
Deletions: 38
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr7 | 98920 | 98921 | 2 | |
| Deletion | Chr4 | 537957 | 537957 | 1 | |
| Deletion | Chr11 | 771017 | 771017 | 1 | |
| Deletion | Chr11 | 1937020 | 1937023 | 4 | LOC_Os11g04570 |
| Deletion | Chr5 | 2071510 | 2071510 | 1 | |
| Deletion | Chr4 | 3805285 | 3805294 | 10 | |
| Deletion | Chr3 | 5924309 | 5924315 | 7 | LOC_Os03g11470 |
| Deletion | Chr11 | 6841327 | 6841344 | 18 | |
| Deletion | Chr5 | 8515160 | 8515161 | 2 | |
| Deletion | Chr10 | 8607955 | 8608118 | 164 | |
| Deletion | Chr5 | 9774143 | 9774143 | 1 | |
| Deletion | Chr10 | 12093511 | 12093518 | 8 | |
| Deletion | Chr5 | 12801639 | 12801640 | 2 | |
| Deletion | Chr6 | 14291110 | 14291115 | 6 | |
| Deletion | Chr7 | 15042203 | 15042247 | 45 | |
| Deletion | Chr12 | 15118620 | 15118621 | 2 | |
| Deletion | Chr2 | 15400075 | 15400081 | 7 | |
| Deletion | Chr8 | 15533155 | 15533155 | 1 | |
| Deletion | Chr5 | 15670693 | 15670696 | 4 | LOC_Os05g26970 |
| Deletion | Chr8 | 15943505 | 15943505 | 1 | |
| Deletion | Chr10 | 16014368 | 16014368 | 1 | |
| Deletion | Chr4 | 16550673 | 16550673 | 1 | |
| Deletion | Chr10 | 16807973 | 16807977 | 5 | LOC_Os10g31990 |
| Deletion | Chr10 | 18031104 | 18031104 | 1 | |
| Deletion | Chr6 | 18938655 | 18938657 | 3 | |
| Deletion | Chr4 | 21379757 | 21379758 | 2 | |
| Deletion | Chr5 | 21419001 | 21498000 | 79000 | 18 |
| Deletion | Chr12 | 21905921 | 21905921 | 1 | |
| Deletion | Chr12 | 22816522 | 22816523 | 2 | |
| Deletion | Chr2 | 23145394 | 23145394 | 1 | |
| Deletion | Chr1 | 23424765 | 23424765 | 1 | |
| Deletion | Chr5 | 25078614 | 25078614 | 1 | |
| Deletion | Chr1 | 27352713 | 27352713 | 1 | |
| Deletion | Chr6 | 28947646 | 28947646 | 1 | |
| Deletion | Chr2 | 31824249 | 31824257 | 9 | LOC_Os02g51970 |
| Deletion | Chr3 | 33417710 | 33417710 | 1 | |
| Deletion | Chr1 | 40192330 | 40192335 | 6 | |
| Deletion | Chr1 | 41620918 | 41620918 | 1 |
Insertions: 21
Inversions: 5
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr8 | 3060194 | 3200560 | LOC_Os08g05930 |
| Inversion | Chr8 | 3060205 | 3200567 | LOC_Os08g05930 |
| Inversion | Chr12 | 4570447 | 4721673 | LOC_Os12g09030 |
| Inversion | Chr12 | 4573582 | 4721679 | LOC_Os12g09030 |
| Inversion | Chr7 | 10815149 | 10815505 |
Translocations: 4
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr5 | 3985869 | Chr2 | 32316953 | |
| Translocation | Chr5 | 3986055 | Chr2 | 32316958 | |
| Translocation | Chr6 | 5355711 | Chr5 | 3296648 | |
| Translocation | Chr6 | 5355721 | Chr5 | 3304521 |
