Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN4597-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN4597-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 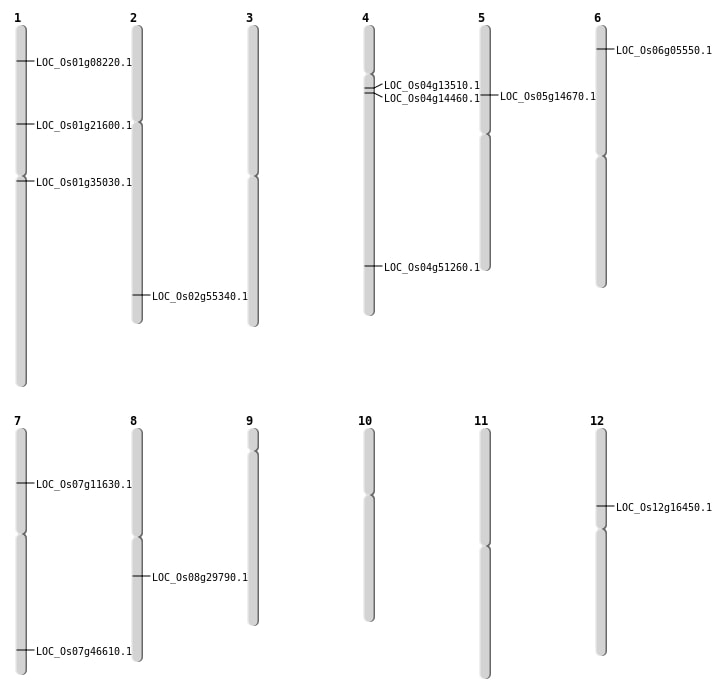
|
| Variant Information |
|---|
Single base substitutions: 35
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 16259071 | T-C | HET | INTERGENIC | |
| SBS | Chr1 | 18305930 | G-A | HET | INTRON | |
| SBS | Chr1 | 23783179 | C-A | HET | INTRON | |
| SBS | Chr1 | 42574351 | T-A | HET | INTERGENIC | |
| SBS | Chr1 | 43269181 | C-T | HET | #VALUE! | |
| SBS | Chr10 | 11678592 | G-T | HET | SYNONYMOUS_CODING | |
| SBS | Chr11 | 26502113 | A-T | HET | INTRON | |
| SBS | Chr12 | 2618738 | G-T | HET | INTERGENIC | |
| SBS | Chr12 | 9422957 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os12g16450 |
| SBS | Chr2 | 12639521 | G-A | HET | INTERGENIC | |
| SBS | Chr2 | 14390626 | A-C | HET | INTERGENIC | |
| SBS | Chr2 | 33888834 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os02g55340 |
| SBS | Chr2 | 7184372 | G-A | HET | SYNONYMOUS_CODING | |
| SBS | Chr2 | 8956476 | T-C | HET | INTRON | |
| SBS | Chr3 | 10504334 | G-A | HOMO | INTERGENIC | |
| SBS | Chr3 | 31147127 | T-G | HET | INTRON | |
| SBS | Chr3 | 5069605 | A-G | HET | INTERGENIC | |
| SBS | Chr3 | 6823451 | C-A | HET | INTERGENIC | |
| SBS | Chr4 | 20223659 | A-G | HET | INTERGENIC | |
| SBS | Chr4 | 23449607 | G-T | HET | INTERGENIC | |
| SBS | Chr4 | 25536855 | C-T | HET | INTRON | |
| SBS | Chr4 | 25536857 | A-G | HET | INTRON | |
| SBS | Chr4 | 30373492 | G-T | HET | NON_SYNONYMOUS_CODING | LOC_Os04g51260 |
| SBS | Chr4 | 32658107 | C-G | HET | SYNONYMOUS_CODING | |
| SBS | Chr4 | 8096351 | C-A | HET | NON_SYNONYMOUS_CODING | LOC_Os04g14460 |
| SBS | Chr4 | 8096352 | C-A | HET | NON_SYNONYMOUS_CODING | LOC_Os04g14460 |
| SBS | Chr5 | 22794492 | C-A | HET | INTRON | |
| SBS | Chr5 | 8340745 | C-T | HOMO | NON_SYNONYMOUS_CODING | LOC_Os05g14670 |
| SBS | Chr6 | 6660754 | C-G | HET | INTERGENIC | |
| SBS | Chr7 | 10650417 | G-A | HET | SYNONYMOUS_CODING | |
| SBS | Chr7 | 2597449 | G-A | HET | INTRON | |
| SBS | Chr7 | 5185455 | C-T | HET | INTERGENIC | |
| SBS | Chr7 | 8230060 | G-A | HET | SYNONYMOUS_CODING | |
| SBS | Chr8 | 18313969 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os08g29790 |
| SBS | Chr9 | 16404469 | C-A | HET | INTRON |
Deletions: 19
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr12 | 2887805 | 2887806 | 1 | |
| Deletion | Chr5 | 3921096 | 3921099 | 3 | |
| Deletion | Chr3 | 4374208 | 4374228 | 20 | |
| Deletion | Chr12 | 6070229 | 6070230 | 1 | |
| Deletion | Chr11 | 6207920 | 6207921 | 1 | |
| Deletion | Chr7 | 7012063 | 7012064 | 1 | |
| Deletion | Chr1 | 7384535 | 7384536 | 1 | |
| Deletion | Chr4 | 7532229 | 7532231 | 2 | LOC_Os04g13510 |
| Deletion | Chr10 | 13441783 | 13441784 | 1 | |
| Deletion | Chr11 | 16553034 | 16553037 | 3 | |
| Deletion | Chr6 | 18138200 | 18138214 | 14 | |
| Deletion | Chr12 | 19386798 | 19386799 | 1 | |
| Deletion | Chr8 | 24072949 | 24072953 | 4 | |
| Deletion | Chr1 | 26553398 | 26553400 | 2 | |
| Deletion | Chr3 | 27218886 | 27218887 | 1 | |
| Deletion | Chr7 | 27847621 | 27847625 | 4 | LOC_Os07g46610 |
| Deletion | Chr2 | 30119658 | 30119659 | 1 | |
| Deletion | Chr2 | 32366699 | 32366705 | 6 | |
| Deletion | Chr3 | 35618340 | 35618358 | 18 |
Insertions: 9
Inversions: 5
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr6 | 2517875 | 2518072 | LOC_Os06g05550 |
| Inversion | Chr1 | 3956765 | 4004506 | LOC_Os01g08220 |
| Inversion | Chr1 | 3956770 | 4004641 | LOC_Os01g08220 |
| Inversion | Chr6 | 19341724 | 19341913 | |
| Inversion | Chr1 | 19385032 | 19881568 | LOC_Os01g35030 |
Translocations: 5
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr2 | 660019 | Chr1 | 27759415 | |
| Translocation | Chr2 | 1012783 | Chr1 | 19385031 | |
| Translocation | Chr8 | 6416545 | Chr7 | 9479733 | LOC_Os07g11630 |
| Translocation | Chr8 | 12102039 | Chr1 | 10160462 | LOC_Os01g21600 |
| Translocation | Chr8 | 12102046 | Chr1 | 10182083 | LOC_Os01g21600 |
