Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN4668-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN4668-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 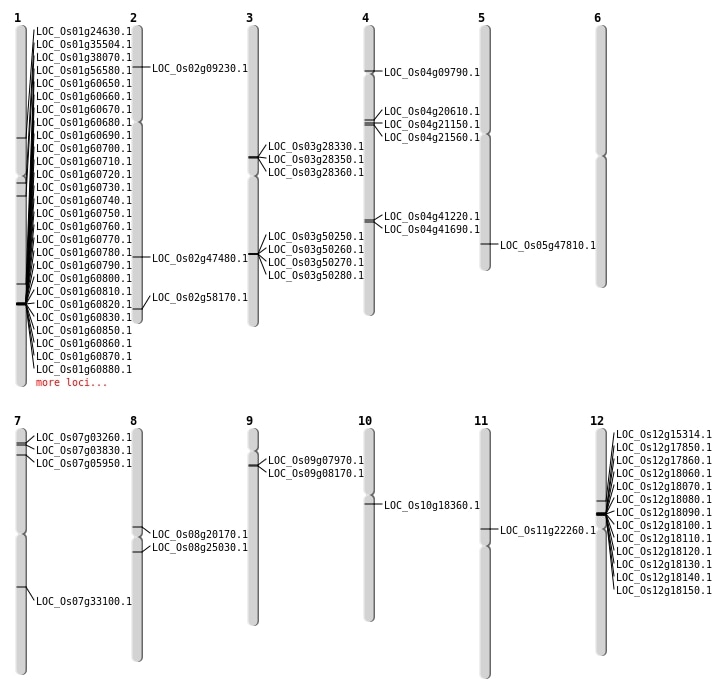
|
| Variant Information |
|---|
Single base substitutions: 46
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 11997752 | G-A | HET | ||
| SBS | Chr1 | 16082508 | C-T | HET | ||
| SBS | Chr1 | 19649944 | G-A | HET | LOC_Os01g35504 | |
| SBS | Chr1 | 24528793 | G-T | HET | ||
| SBS | Chr1 | 24528794 | C-T | HET | ||
| SBS | Chr1 | 43140227 | A-G | HOMO | ||
| SBS | Chr1 | 9088708 | C-A | HOMO | ||
| SBS | Chr10 | 5530649 | C-G | HOMO | ||
| SBS | Chr10 | 6670559 | G-T | HET | ||
| SBS | Chr10 | 8023468 | T-C | HET | ||
| SBS | Chr11 | 9401229 | C-T | HET | ||
| SBS | Chr12 | 16907798 | A-T | HET | ||
| SBS | Chr2 | 12776055 | C-T | HOMO | ||
| SBS | Chr2 | 14469978 | G-T | HET | ||
| SBS | Chr2 | 26048495 | A-G | HET | ||
| SBS | Chr2 | 29006669 | G-A | HET | LOC_Os02g47480 | |
| SBS | Chr2 | 30841087 | C-G | HET | ||
| SBS | Chr2 | 6744115 | A-G | HET | ||
| SBS | Chr2 | 9351480 | C-G | HET | ||
| SBS | Chr3 | 20781834 | A-G | HET | ||
| SBS | Chr3 | 21694930 | G-A | HET | ||
| SBS | Chr4 | 10347369 | G-A | HET | ||
| SBS | Chr4 | 11906748 | C-T | HET | LOC_Os04g21150 | |
| SBS | Chr4 | 22469191 | C-T | HET | ||
| SBS | Chr4 | 23239996 | C-A | HET | ||
| SBS | Chr4 | 25573787 | C-A | HET | ||
| SBS | Chr4 | 27414593 | C-T | HET | ||
| SBS | Chr4 | 32212978 | C-T | HET | ||
| SBS | Chr4 | 368295 | A-G | HOMO | ||
| SBS | Chr5 | 16862392 | C-T | HOMO | ||
| SBS | Chr5 | 19244169 | A-T | HET | ||
| SBS | Chr5 | 21076856 | C-T | HET | ||
| SBS | Chr5 | 23642995 | G-T | HET | ||
| SBS | Chr5 | 23845346 | G-T | HET | ||
| SBS | Chr5 | 2952793 | T-C | HOMO | ||
| SBS | Chr6 | 25217570 | G-A | HET | ||
| SBS | Chr6 | 4256403 | G-A | HOMO | ||
| SBS | Chr7 | 10404487 | C-T | HET | ||
| SBS | Chr7 | 18387717 | C-T | HET | ||
| SBS | Chr7 | 26939431 | C-T | HET | ||
| SBS | Chr7 | 27555883 | A-C | HET | ||
| SBS | Chr7 | 28369899 | T-C | HET | ||
| SBS | Chr8 | 12088753 | G-A | HOMO | LOC_Os08g20170 | |
| SBS | Chr8 | 12088754 | C-A | HOMO | ||
| SBS | Chr8 | 12088755 | T-A | HOMO | LOC_Os08g20170 | |
| SBS | Chr9 | 21840101 | G-A | HET |
Deletions: 28
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr6 | 77060 | 77060 | 1 | |
| Deletion | Chr9 | 298544 | 298545 | 2 | |
| Deletion | Chr4 | 1528715 | 1528722 | 8 | |
| Deletion | Chr9 | 1938415 | 1938465 | 51 | |
| Deletion | Chr3 | 3237018 | 3237018 | 1 | |
| Deletion | Chr9 | 4234246 | 4234252 | 7 | LOC_Os09g08170 |
| Deletion | Chr4 | 6378642 | 6378685 | 44 | |
| Deletion | Chr4 | 6777027 | 6777027 | 1 | |
| Deletion | Chr12 | 8727722 | 8727725 | 4 | LOC_Os12g15314 |
| Deletion | Chr2 | 9207081 | 9207081 | 1 | |
| Deletion | Chr10 | 9326822 | 9326824 | 3 | LOC_Os10g18360 |
| Deletion | Chr6 | 9340933 | 9340934 | 2 | |
| Deletion | Chr1 | 9353794 | 9353797 | 4 | |
| Deletion | Chr12 | 10225001 | 10238000 | 13000 | 2 |
| Deletion | Chr12 | 10386001 | 10481000 | 95000 | 10 |
| Deletion | Chr8 | 11541515 | 11541515 | 1 | |
| Deletion | Chr10 | 13750055 | 13750060 | 6 | |
| Deletion | Chr1 | 13873512 | 13873512 | 1 | LOC_Os01g24630 |
| Deletion | Chr3 | 16307001 | 16344000 | 37000 | 3 |
| Deletion | Chr6 | 19060644 | 19060644 | 1 | |
| Deletion | Chr4 | 24713489 | 24713489 | 1 | LOC_Os04g41690 |
| Deletion | Chr12 | 27036813 | 27036814 | 2 | |
| Deletion | Chr3 | 28647001 | 28660000 | 13000 | 4 |
| Deletion | Chr5 | 29701218 | 29701218 | 1 | |
| Deletion | Chr1 | 35066001 | 35114000 | 48000 | 7 |
| Deletion | Chr1 | 35119001 | 35272000 | 153000 | 23 |
| Deletion | Chr1 | 35285001 | 35324000 | 39000 | 7 |
| Deletion | Chr1 | 35326001 | 35354000 | 28000 | 5 |
Insertions: 21
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr1 | 21329849 | 21329849 | 1 | LOC_Os01g38070 |
| Insertion | Chr11 | 20216643 | 20216644 | 2 | |
| Insertion | Chr11 | 25271686 | 25271686 | 1 | |
| Insertion | Chr2 | 8773389 | 8773389 | 1 | |
| Insertion | Chr3 | 25905419 | 25905515 | 97 | |
| Insertion | Chr3 | 33993649 | 33993649 | 1 | |
| Insertion | Chr3 | 5540969 | 5540969 | 1 | |
| Insertion | Chr3 | 5845687 | 5845687 | 1 | |
| Insertion | Chr4 | 10337162 | 10337162 | 1 | |
| Insertion | Chr4 | 18043642 | 18043642 | 1 | |
| Insertion | Chr4 | 24456019 | 24456057 | 39 | LOC_Os04g41220 |
| Insertion | Chr4 | 9227864 | 9227864 | 1 | |
| Insertion | Chr5 | 1982410 | 1982417 | 8 | |
| Insertion | Chr6 | 18652730 | 18652730 | 1 | |
| Insertion | Chr6 | 9968314 | 9968314 | 1 | |
| Insertion | Chr7 | 2382367 | 2382367 | 1 | |
| Insertion | Chr7 | 26463402 | 26463403 | 2 | |
| Insertion | Chr7 | 28627275 | 28627275 | 1 | |
| Insertion | Chr8 | 15197944 | 15197955 | 12 | LOC_Os08g25030 |
| Insertion | Chr8 | 4767461 | 4767462 | 2 | |
| Insertion | Chr9 | 7373172 | 7373218 | 47 |
Inversions: 4
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr7 | 1305619 | 1600648 | 2 |
| Inversion | Chr4 | 11534209 | 12186494 | 2 |
| Inversion | Chr2 | 34840853 | 35673892 | |
| Inversion | Chr2 | 34840860 | 35673900 |
Translocations: 7
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr7 | 1305610 | Chr3 | 16359956 | LOC_Os07g03260 |
| Translocation | Chr7 | 1600647 | Chr1 | 32620013 | 2 |
| Translocation | Chr7 | 2882524 | Chr3 | 16360511 | LOC_Os07g05950 |
| Translocation | Chr7 | 2882532 | Chr1 | 32620005 | 2 |
| Translocation | Chr7 | 2882873 | Chr3 | 16344535 | LOC_Os07g05950 |
| Translocation | Chr7 | 2882880 | Chr3 | 16359980 | LOC_Os07g05950 |
| Translocation | Chr12 | 8728445 | Chr1 | 35371274 |
