Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN4680-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN4680-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 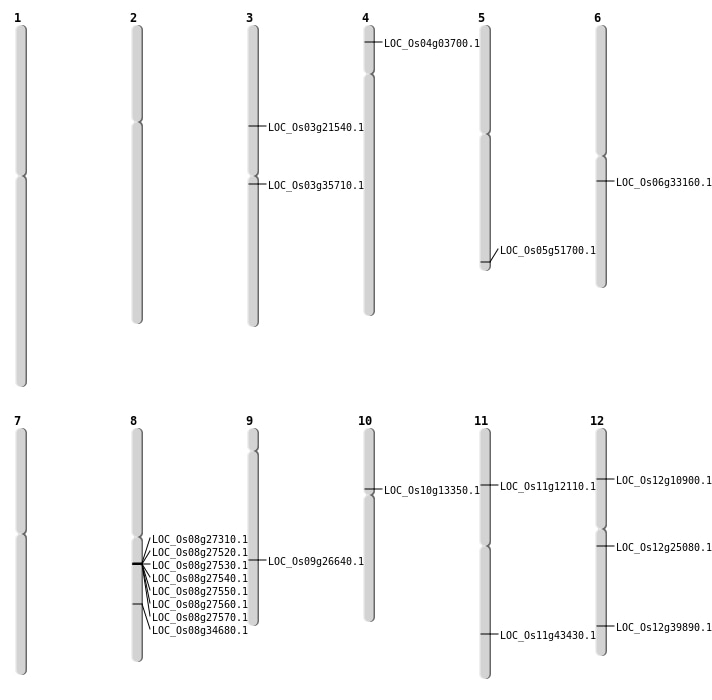
|
| Variant Information |
|---|
Single base substitutions: 44
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 14593186 | C-A | HET | ||
| SBS | Chr1 | 21079068 | C-A | HET | ||
| SBS | Chr1 | 5382667 | T-C | HOMO | ||
| SBS | Chr10 | 13767734 | C-T | HET | ||
| SBS | Chr10 | 21159811 | G-C | HET | ||
| SBS | Chr10 | 7209217 | G-C | HET | LOC_Os10g13350 | |
| SBS | Chr11 | 21422111 | C-T | HET | ||
| SBS | Chr11 | 26876363 | G-A | HET | ||
| SBS | Chr12 | 10844427 | G-A | HET | ||
| SBS | Chr12 | 13968897 | G-T | HET | ||
| SBS | Chr12 | 14397075 | C-A | HET | ||
| SBS | Chr12 | 14408991 | A-T | HET | LOC_Os12g25080 | |
| SBS | Chr12 | 21722005 | A-C | HET | ||
| SBS | Chr12 | 24650168 | C-A | HET | LOC_Os12g39890 | |
| SBS | Chr2 | 11243101 | T-C | HOMO | ||
| SBS | Chr2 | 31207099 | C-A | HET | ||
| SBS | Chr2 | 694338 | G-A | HET | ||
| SBS | Chr2 | 8919138 | A-G | HOMO | ||
| SBS | Chr3 | 16568930 | T-C | HET | ||
| SBS | Chr3 | 22843833 | C-A | HOMO | ||
| SBS | Chr3 | 27381024 | T-A | HET | ||
| SBS | Chr3 | 33122697 | C-T | HOMO | ||
| SBS | Chr4 | 11354607 | T-G | HET | ||
| SBS | Chr4 | 14021597 | T-G | HOMO | ||
| SBS | Chr4 | 26826213 | A-G | HOMO | ||
| SBS | Chr4 | 3212445 | G-A | HET | ||
| SBS | Chr4 | 9931721 | A-G | HET | ||
| SBS | Chr5 | 27852405 | G-A | HOMO | ||
| SBS | Chr6 | 13370616 | G-A | HET | ||
| SBS | Chr6 | 18581 | T-A | HOMO | ||
| SBS | Chr6 | 19945797 | G-T | HOMO | ||
| SBS | Chr6 | 25012402 | C-T | HET | ||
| SBS | Chr7 | 10117367 | A-G | HET | ||
| SBS | Chr7 | 10608673 | C-T | HOMO | ||
| SBS | Chr7 | 12562800 | A-G | HET | ||
| SBS | Chr7 | 1883994 | G-A | HOMO | ||
| SBS | Chr7 | 27305421 | A-T | HET | ||
| SBS | Chr7 | 28226866 | T-A | HET | ||
| SBS | Chr7 | 4047563 | G-A | HOMO | ||
| SBS | Chr7 | 7359651 | T-C | HET | ||
| SBS | Chr8 | 12761748 | G-A | HET | ||
| SBS | Chr8 | 23179984 | G-A | HET | ||
| SBS | Chr8 | 26172160 | G-T | HET | ||
| SBS | Chr8 | 8952951 | T-C | HET |
Deletions: 44
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr6 | 671892 | 671904 | 13 | |
| Deletion | Chr4 | 855444 | 855444 | 1 | |
| Deletion | Chr3 | 1038339 | 1038339 | 1 | |
| Deletion | Chr4 | 1633937 | 1633946 | 10 | LOC_Os04g03700 |
| Deletion | Chr12 | 2127317 | 2127317 | 1 | |
| Deletion | Chr4 | 2617320 | 2617329 | 10 | |
| Deletion | Chr2 | 3590576 | 3590576 | 1 | |
| Deletion | Chr2 | 3818137 | 3818137 | 1 | |
| Deletion | Chr4 | 4206030 | 4206030 | 1 | |
| Deletion | Chr3 | 5700814 | 5700814 | 1 | |
| Deletion | Chr9 | 5853478 | 5853488 | 11 | |
| Deletion | Chr11 | 7140700 | 7140700 | 1 | |
| Deletion | Chr8 | 7384957 | 7384973 | 17 | |
| Deletion | Chr2 | 9121091 | 9121136 | 46 | |
| Deletion | Chr9 | 10508594 | 10508594 | 1 | |
| Deletion | Chr3 | 10795077 | 10795077 | 1 | |
| Deletion | Chr4 | 11102479 | 11102482 | 4 | |
| Deletion | Chr12 | 12048958 | 12048958 | 1 | |
| Deletion | Chr3 | 12310960 | 12310963 | 4 | LOC_Os03g21540 |
| Deletion | Chr4 | 13158291 | 13158291 | 1 | |
| Deletion | Chr6 | 13355946 | 13355946 | 1 | |
| Deletion | Chr4 | 14066295 | 14066295 | 1 | |
| Deletion | Chr2 | 14769713 | 14769716 | 4 | |
| Deletion | Chr11 | 15784365 | 15784365 | 1 | |
| Deletion | Chr9 | 16162556 | 16162620 | 65 | LOC_Os09g26640 |
| Deletion | Chr3 | 16374547 | 16374547 | 1 | |
| Deletion | Chr8 | 16789001 | 16806000 | 17000 | 6 |
| Deletion | Chr4 | 17425601 | 17425604 | 4 | |
| Deletion | Chr7 | 17560456 | 17560478 | 23 | |
| Deletion | Chr6 | 18408281 | 18408354 | 74 | |
| Deletion | Chr6 | 18706664 | 18706664 | 1 | |
| Deletion | Chr8 | 21178244 | 21178255 | 12 | |
| Deletion | Chr9 | 21831559 | 21831577 | 19 | |
| Deletion | Chr12 | 22852232 | 22852236 | 5 | |
| Deletion | Chr2 | 22886718 | 22886718 | 1 | |
| Deletion | Chr1 | 23508269 | 23508311 | 43 | |
| Deletion | Chr2 | 23726362 | 23726364 | 3 | |
| Deletion | Chr1 | 24037825 | 24037832 | 8 | |
| Deletion | Chr4 | 25193238 | 25193247 | 10 | |
| Deletion | Chr1 | 25334606 | 25334621 | 16 | |
| Deletion | Chr4 | 26266087 | 26266103 | 17 | |
| Deletion | Chr8 | 27504852 | 27504853 | 2 | |
| Deletion | Chr2 | 27740346 | 27740381 | 36 | |
| Deletion | Chr5 | 29663237 | 29663245 | 9 | LOC_Os05g51700 |
Insertions: 10
Inversions: 8
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr8 | 7796853 | 7797145 | |
| Inversion | Chr1 | 11163197 | 11163269 | |
| Inversion | Chr1 | 12500982 | 12501131 | |
| Inversion | Chr8 | 21784295 | 21813614 | LOC_Os08g34680 |
| Inversion | Chr8 | 21784483 | 21812780 | |
| Inversion | Chr1 | 23719734 | 23881188 | |
| Inversion | Chr1 | 23719748 | 23881192 | |
| Inversion | Chr11 | 25188550 | 25188723 |
Translocations: 3
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr6 | 13912855 | Chr3 | 19779915 | LOC_Os03g35710 |
| Translocation | Chr6 | 13912867 | Chr3 | 19772097 | |
| Translocation | Chr12 | 18092743 | Chr3 | 25788773 |
