Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN4688-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN4688-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 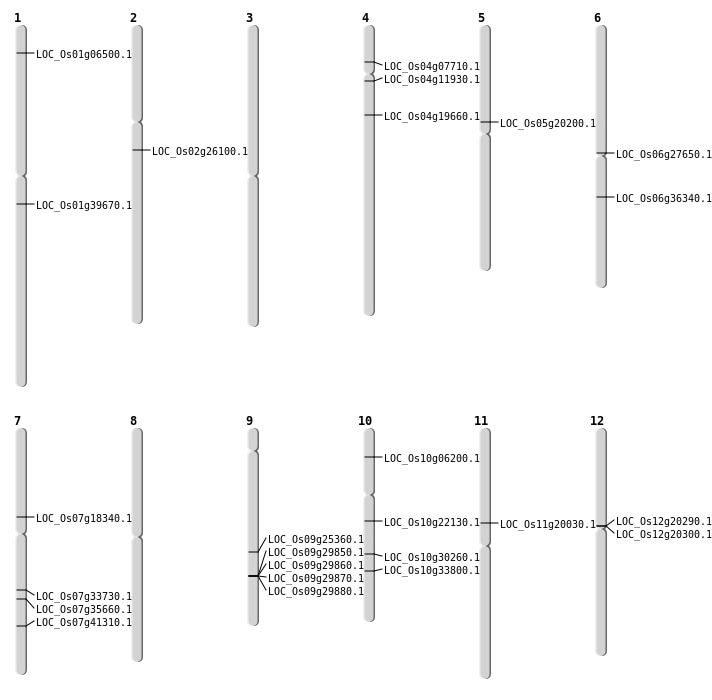
|
| Variant Information |
|---|
Single base substitutions: 63
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 11430158 | C-A | HET | ||
| SBS | Chr1 | 11496910 | T-G | HET | ||
| SBS | Chr1 | 16938974 | C-A | HET | ||
| SBS | Chr1 | 22377036 | A-G | HET | LOC_Os01g39670 | |
| SBS | Chr1 | 22978877 | C-T | HET | ||
| SBS | Chr1 | 2677769 | T-A | HOMO | ||
| SBS | Chr1 | 3059177 | T-A | HOMO | LOC_Os01g06500 | |
| SBS | Chr1 | 33381907 | G-T | HET | ||
| SBS | Chr1 | 40767526 | T-A | HOMO | ||
| SBS | Chr1 | 5180148 | C-T | HET | ||
| SBS | Chr1 | 9782448 | T-C | HET | ||
| SBS | Chr10 | 11432889 | G-T | HET | LOC_Os10g22130 | |
| SBS | Chr10 | 17917168 | G-A | HET | LOC_Os10g33800 | |
| SBS | Chr10 | 17917169 | T-A | HET | ||
| SBS | Chr10 | 4928722 | C-A | HET | ||
| SBS | Chr12 | 14422628 | T-A | HET | ||
| SBS | Chr12 | 14422629 | A-G | HET | ||
| SBS | Chr12 | 15360403 | G-T | HET | ||
| SBS | Chr12 | 2325089 | G-T | HET | ||
| SBS | Chr2 | 10966132 | T-A | HET | ||
| SBS | Chr2 | 11637663 | C-T | HET | ||
| SBS | Chr2 | 14560358 | C-T | HET | ||
| SBS | Chr2 | 15336426 | A-T | HOMO | LOC_Os02g26100 | |
| SBS | Chr2 | 18874810 | T-C | HET | ||
| SBS | Chr2 | 20637183 | T-C | HET | ||
| SBS | Chr2 | 33186571 | G-A | HET | ||
| SBS | Chr3 | 11688708 | G-A | HOMO | ||
| SBS | Chr3 | 17346457 | G-T | HET | ||
| SBS | Chr3 | 17712956 | C-T | HET | ||
| SBS | Chr3 | 22843868 | G-A | HET | ||
| SBS | Chr3 | 24631136 | A-G | HET | ||
| SBS | Chr3 | 24649969 | G-A | HET | ||
| SBS | Chr4 | 11942064 | G-A | HET | ||
| SBS | Chr4 | 17220807 | G-T | HET | ||
| SBS | Chr4 | 20443476 | G-T | HET | ||
| SBS | Chr4 | 6402381 | G-A | HET | ||
| SBS | Chr4 | 6547665 | G-T | HET | LOC_Os04g11930 | |
| SBS | Chr4 | 7679161 | A-G | HOMO | ||
| SBS | Chr5 | 11823805 | C-G | HET | LOC_Os05g20200 | |
| SBS | Chr5 | 13597985 | C-T | HET | ||
| SBS | Chr5 | 14049756 | A-C | HET | ||
| SBS | Chr5 | 22561471 | G-T | HET | ||
| SBS | Chr5 | 7997782 | T-C | HET | ||
| SBS | Chr5 | 9422521 | G-A | HET | ||
| SBS | Chr6 | 17399795 | G-A | HET | ||
| SBS | Chr6 | 27367974 | C-T | HET | ||
| SBS | Chr6 | 28184923 | A-T | HET | ||
| SBS | Chr6 | 29640649 | T-C | HET | ||
| SBS | Chr6 | 4800082 | C-A | HET | ||
| SBS | Chr7 | 10866548 | T-A | HET | LOC_Os07g18340 | |
| SBS | Chr7 | 21636124 | C-T | HET | ||
| SBS | Chr7 | 28839536 | C-A | HET | ||
| SBS | Chr7 | 4126130 | G-A | HET | ||
| SBS | Chr8 | 11081161 | T-G | HET | ||
| SBS | Chr8 | 12284292 | A-T | HET | ||
| SBS | Chr8 | 16480049 | G-A | HET | ||
| SBS | Chr8 | 17101861 | T-C | HET | ||
| SBS | Chr8 | 20650920 | T-A | HET | ||
| SBS | Chr8 | 7182081 | T-A | HET | ||
| SBS | Chr9 | 16453389 | A-G | HET | ||
| SBS | Chr9 | 19355230 | T-C | HET | ||
| SBS | Chr9 | 5898641 | G-A | HET | ||
| SBS | Chr9 | 8636531 | C-A | HET |
Deletions: 31
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr5 | 2232935 | 2232939 | 5 | |
| Deletion | Chr12 | 2458904 | 2458904 | 1 | |
| Deletion | Chr8 | 2701566 | 2701566 | 1 | |
| Deletion | Chr1 | 3041960 | 3041963 | 4 | |
| Deletion | Chr10 | 3177911 | 3177912 | 2 | LOC_Os10g06200 |
| Deletion | Chr3 | 3258395 | 3258400 | 6 | |
| Deletion | Chr9 | 6350747 | 6350747 | 1 | |
| Deletion | Chr5 | 10096998 | 10096999 | 2 | |
| Deletion | Chr12 | 11413331 | 11413333 | 3 | |
| Deletion | Chr11 | 11526809 | 11526856 | 48 | LOC_Os11g20030 |
| Deletion | Chr12 | 11847001 | 11857000 | 10000 | 2 |
| Deletion | Chr11 | 12093572 | 12093574 | 3 | |
| Deletion | Chr12 | 12818824 | 12818824 | 1 | |
| Deletion | Chr7 | 13789093 | 13789093 | 1 | |
| Deletion | Chr11 | 13948555 | 13948559 | 5 | |
| Deletion | Chr6 | 14857211 | 14857211 | 1 | |
| Deletion | Chr9 | 15339665 | 15339673 | 9 | |
| Deletion | Chr6 | 15653526 | 15653532 | 7 | LOC_Os06g27650 |
| Deletion | Chr5 | 16609649 | 16609649 | 1 | |
| Deletion | Chr5 | 17854674 | 17854681 | 8 | |
| Deletion | Chr9 | 18156001 | 18173000 | 17000 | 4 |
| Deletion | Chr4 | 18372685 | 18372685 | 1 | |
| Deletion | Chr3 | 20026338 | 20026338 | 1 | |
| Deletion | Chr10 | 20502589 | 20502589 | 1 | |
| Deletion | Chr2 | 21938220 | 21938220 | 1 | |
| Deletion | Chr7 | 23691906 | 23691906 | 1 | |
| Deletion | Chr7 | 23711029 | 23711036 | 8 | |
| Deletion | Chr6 | 26413143 | 26413143 | 1 | |
| Deletion | Chr2 | 27828843 | 27828850 | 8 | |
| Deletion | Chr2 | 29633989 | 29633990 | 2 | |
| Deletion | Chr6 | 30292873 | 30292873 | 1 |
Insertions: 17
Inversions: 3
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr10 | 15722579 | 15903171 | LOC_Os10g30260 |
| Inversion | Chr7 | 21360301 | 21386461 | LOC_Os07g35660 |
| Inversion | Chr7 | 21360303 | 21386459 | LOC_Os07g35660 |
Translocations: 5
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr9 | 6164867 | Chr4 | 10967289 | LOC_Os04g19660 |
| Translocation | Chr11 | 18880801 | Chr9 | 6164869 | |
| Translocation | Chr11 | 18881090 | Chr4 | 10966891 | LOC_Os04g19660 |
| Translocation | Chr8 | 26329658 | Chr7 | 17504014 | |
| Translocation | Chr8 | 26331937 | Chr7 | 17504021 |
