Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN4752-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN4752-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 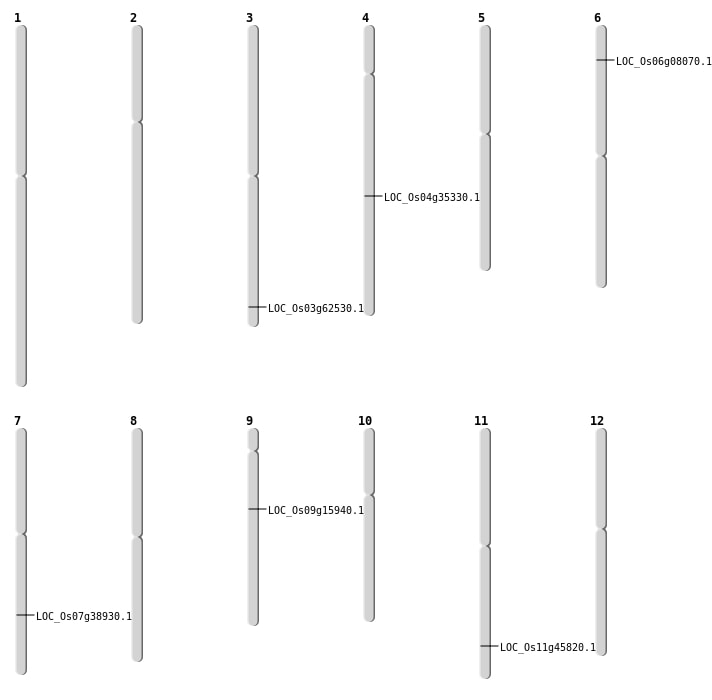
|
| Variant Information |
|---|
Single base substitutions: 46
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 11469748 | C-T | HET | INTERGENIC | |
| SBS | Chr1 | 14535461 | G-T | HET | INTERGENIC | |
| SBS | Chr1 | 42469810 | G-C | HET | INTERGENIC | |
| SBS | Chr11 | 11087720 | C-T | HET | INTERGENIC | |
| SBS | Chr11 | 24871543 | G-A | HOMO | SYNONYMOUS_CODING | |
| SBS | Chr11 | 25268784 | C-A | HET | INTERGENIC | |
| SBS | Chr12 | 14317427 | G-T | HOMO | INTERGENIC | |
| SBS | Chr12 | 2000252 | G-A | HET | INTERGENIC | |
| SBS | Chr12 | 25762110 | T-C | HET | INTERGENIC | |
| SBS | Chr12 | 8600992 | C-G | HET | INTRON | |
| SBS | Chr12 | 9864108 | C-A | HET | INTERGENIC | |
| SBS | Chr2 | 3439387 | A-G | HET | INTERGENIC | |
| SBS | Chr2 | 35105666 | C-A | HET | INTERGENIC | |
| SBS | Chr2 | 8249448 | C-A | HET | INTERGENIC | |
| SBS | Chr3 | 1373090 | C-A | HET | SYNONYMOUS_CODING | |
| SBS | Chr3 | 18858162 | T-G | HET | INTERGENIC | |
| SBS | Chr3 | 25577455 | C-T | HOMO | SYNONYMOUS_CODING | |
| SBS | Chr3 | 35408195 | C-G | HET | SYNONYMOUS_CODING | |
| SBS | Chr3 | 35408197 | A-T | HET | NON_SYNONYMOUS_CODING | LOC_Os03g62530 |
| SBS | Chr3 | 9586779 | C-T | HET | INTERGENIC | |
| SBS | Chr5 | 15902665 | G-A | HET | INTERGENIC | |
| SBS | Chr5 | 16607199 | A-T | HET | INTERGENIC | |
| SBS | Chr5 | 22727572 | A-C | HET | INTRON | |
| SBS | Chr5 | 23359971 | G-T | HET | SYNONYMOUS_CODING | |
| SBS | Chr5 | 23936365 | C-T | HET | INTRON | |
| SBS | Chr5 | 24084797 | C-A | HET | SPLICE_SITE_REGION | |
| SBS | Chr5 | 6382614 | T-A | HET | INTERGENIC | |
| SBS | Chr5 | 8000642 | T-A | HET | UTR_5_PRIME | |
| SBS | Chr6 | 12569033 | C-A | HOMO | INTERGENIC | |
| SBS | Chr6 | 28364582 | G-A | HOMO | INTRON | |
| SBS | Chr6 | 30792945 | T-A | HOMO | INTERGENIC | |
| SBS | Chr6 | 3905306 | T-A | HET | NON_SYNONYMOUS_CODING | LOC_Os06g08070 |
| SBS | Chr6 | 3905307 | G-A | HET | SYNONYMOUS_CODING | |
| SBS | Chr6 | 4006809 | A-T | HET | INTERGENIC | |
| SBS | Chr7 | 4419218 | G-A | HET | INTERGENIC | |
| SBS | Chr7 | 4902370 | T-C | HET | INTERGENIC | |
| SBS | Chr7 | 7907112 | A-T | HET | INTERGENIC | |
| SBS | Chr8 | 15263070 | G-A | HET | INTRON | |
| SBS | Chr8 | 19403044 | G-C | HOMO | INTERGENIC | |
| SBS | Chr8 | 2316747 | C-T | HOMO | INTERGENIC | |
| SBS | Chr8 | 23560918 | C-G | HET | INTERGENIC | |
| SBS | Chr8 | 26025794 | C-T | HOMO | INTRON | |
| SBS | Chr9 | 14013217 | G-A | HET | INTERGENIC | |
| SBS | Chr9 | 8962253 | G-T | HET | INTERGENIC | |
| SBS | Chr9 | 9742124 | G-T | HET | NON_SYNONYMOUS_CODING | LOC_Os09g15940 |
| SBS | Chr9 | 9854998 | G-A | HET | INTERGENIC |
Deletions: 22
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr12 | 449284 | 449285 | 1 | |
| Deletion | Chr9 | 945960 | 945961 | 1 | |
| Deletion | Chr9 | 968818 | 968823 | 5 | |
| Deletion | Chr2 | 1707339 | 1707341 | 2 | |
| Deletion | Chr4 | 2621797 | 2621799 | 2 | |
| Deletion | Chr10 | 5355210 | 5355211 | 1 | |
| Deletion | Chr9 | 5425176 | 5425220 | 44 | |
| Deletion | Chr7 | 7926786 | 7926787 | 1 | |
| Deletion | Chr9 | 9132033 | 9132111 | 78 | |
| Deletion | Chr10 | 9497779 | 9497780 | 1 | |
| Deletion | Chr3 | 11654230 | 11654239 | 9 | |
| Deletion | Chr5 | 14482993 | 14482994 | 1 | |
| Deletion | Chr7 | 15072965 | 15072966 | 1 | |
| Deletion | Chr4 | 15938609 | 15938612 | 3 | |
| Deletion | Chr4 | 17481747 | 17481753 | 6 | |
| Deletion | Chr10 | 17562449 | 17562450 | 1 | |
| Deletion | Chr10 | 19648219 | 19648220 | 1 | |
| Deletion | Chr6 | 20801571 | 20801572 | 1 | |
| Deletion | Chr10 | 20843824 | 20843831 | 7 | |
| Deletion | Chr4 | 21486830 | 21486836 | 6 | LOC_Os04g35330 |
| Deletion | Chr8 | 22070538 | 22070540 | 2 | |
| Deletion | Chr12 | 24546733 | 24546736 | 3 |
Insertions: 20
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr1 | 27711602 | 27711654 | 53 | |
| Insertion | Chr1 | 32519431 | 32519431 | 1 | |
| Insertion | Chr11 | 23375812 | 23375814 | 3 | |
| Insertion | Chr11 | 27725711 | 27725754 | 44 | LOC_Os11g45820 |
| Insertion | Chr2 | 2106504 | 2106505 | 2 | |
| Insertion | Chr2 | 6373295 | 6373295 | 1 | |
| Insertion | Chr3 | 13200871 | 13200871 | 1 | |
| Insertion | Chr3 | 61102 | 61103 | 2 | |
| Insertion | Chr4 | 12901740 | 12901782 | 43 | |
| Insertion | Chr4 | 16136278 | 16136278 | 1 | |
| Insertion | Chr4 | 17193021 | 17193021 | 1 | |
| Insertion | Chr4 | 21404855 | 21404855 | 1 | |
| Insertion | Chr4 | 31709213 | 31709214 | 2 | |
| Insertion | Chr5 | 19288873 | 19288873 | 1 | |
| Insertion | Chr5 | 23890190 | 23890190 | 1 | |
| Insertion | Chr5 | 27453926 | 27453926 | 1 | |
| Insertion | Chr8 | 13805227 | 13805227 | 1 | |
| Insertion | Chr8 | 5207523 | 5207524 | 2 | |
| Insertion | Chr9 | 2960970 | 2960971 | 2 | |
| Insertion | Chr9 | 8280003 | 8280003 | 1 |
Inversions: 1
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr7 | 23345160 | 23345244 | LOC_Os07g38930 |
Translocations: 3
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr5 | 14541374 | Chr2 | 25966150 | |
| Translocation | Chr11 | 17693016 | Chr2 | 5950580 | |
| Translocation | Chr6 | 25881613 | Chr2 | 10193788 |
