Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN4758-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN4758-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 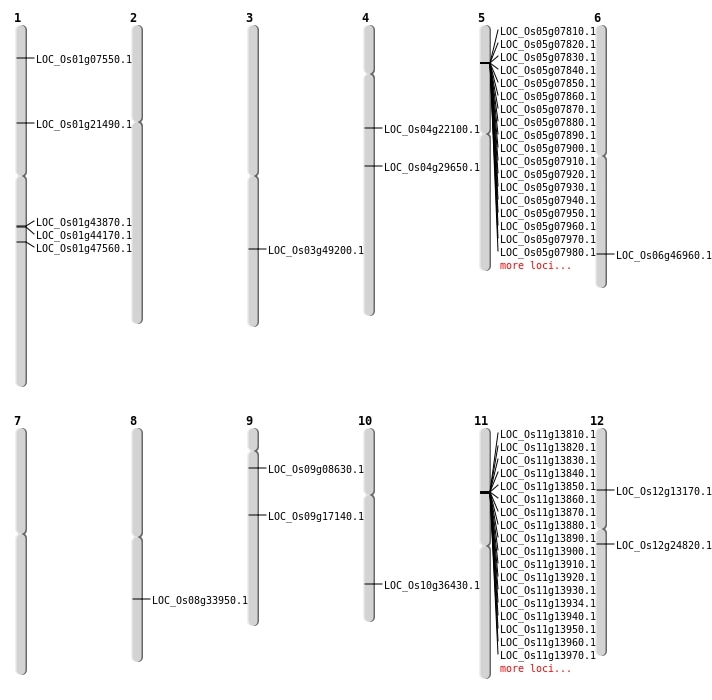
|
| Variant Information |
|---|
Single base substitutions: 74
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 11752203 | C-A | HOMO | ||
| SBS | Chr1 | 20409924 | A-G | HOMO | ||
| SBS | Chr1 | 27199896 | C-T | HET | LOC_Os01g47560 | |
| SBS | Chr1 | 31759789 | G-A | HET | ||
| SBS | Chr1 | 3617099 | C-T | HET | LOC_Os01g07550 | |
| SBS | Chr1 | 3905569 | T-A | HOMO | ||
| SBS | Chr1 | 9493305 | C-T | HET | ||
| SBS | Chr10 | 19493875 | C-T | HET | LOC_Os10g36430 | |
| SBS | Chr10 | 20431621 | A-T | HET | ||
| SBS | Chr10 | 3503992 | G-A | HET | ||
| SBS | Chr10 | 3799063 | A-T | HET | ||
| SBS | Chr10 | 5628132 | C-T | HET | ||
| SBS | Chr11 | 15220400 | C-T | HET | ||
| SBS | Chr11 | 16760171 | G-A | HET | ||
| SBS | Chr11 | 25311462 | C-T | HOMO | ||
| SBS | Chr11 | 25484041 | G-A | HOMO | LOC_Os11g42320 | |
| SBS | Chr11 | 3075001 | C-T | HET | ||
| SBS | Chr11 | 8823449 | C-T | HET | ||
| SBS | Chr11 | 9541791 | G-A | HET | ||
| SBS | Chr12 | 11842866 | G-A | HET | ||
| SBS | Chr12 | 13551380 | T-A | HET | ||
| SBS | Chr12 | 13551381 | T-A | HET | ||
| SBS | Chr12 | 14273485 | C-T | HET | ||
| SBS | Chr12 | 14777503 | A-G | HET | ||
| SBS | Chr12 | 20686761 | G-A | HET | ||
| SBS | Chr12 | 26985804 | C-T | HET | ||
| SBS | Chr2 | 19478153 | T-A | HET | ||
| SBS | Chr2 | 20493358 | A-T | HET | ||
| SBS | Chr2 | 30968402 | C-A | HET | ||
| SBS | Chr2 | 31454235 | T-C | HET | ||
| SBS | Chr3 | 11205504 | G-A | HET | ||
| SBS | Chr3 | 13939450 | C-A | HET | ||
| SBS | Chr3 | 19788113 | A-T | HET | ||
| SBS | Chr3 | 24490428 | G-A | HET | ||
| SBS | Chr3 | 28019409 | A-G | HET | ||
| SBS | Chr3 | 34004044 | C-A | HET | ||
| SBS | Chr4 | 12521024 | C-A | HET | LOC_Os04g22100 | |
| SBS | Chr4 | 16161856 | C-T | HET | ||
| SBS | Chr4 | 16944905 | G-A | HET | ||
| SBS | Chr4 | 17648547 | T-C | HOMO | LOC_Os04g29650 | |
| SBS | Chr4 | 17888899 | G-A | HOMO | ||
| SBS | Chr4 | 1811410 | C-T | HET | ||
| SBS | Chr4 | 1811414 | G-T | HET | ||
| SBS | Chr4 | 21057798 | G-A | HOMO | ||
| SBS | Chr4 | 21905692 | T-A | HOMO | ||
| SBS | Chr4 | 23225055 | G-T | HOMO | ||
| SBS | Chr4 | 34964977 | T-C | HET | ||
| SBS | Chr4 | 8328965 | C-A | HET | ||
| SBS | Chr4 | 9873615 | C-A | HET | ||
| SBS | Chr5 | 11687385 | C-T | HET | LOC_Os05g19990 | |
| SBS | Chr5 | 21600189 | G-T | HET | ||
| SBS | Chr5 | 21621562 | C-T | HET | ||
| SBS | Chr5 | 24143510 | G-T | HET | ||
| SBS | Chr5 | 2681614 | G-A | HET | ||
| SBS | Chr5 | 3302259 | T-A | HET | ||
| SBS | Chr5 | 6202477 | T-A | HET | ||
| SBS | Chr5 | 7365611 | G-T | HET | ||
| SBS | Chr5 | 9541120 | T-A | HET | ||
| SBS | Chr6 | 14270898 | C-T | HOMO | ||
| SBS | Chr6 | 20822813 | A-G | HET | ||
| SBS | Chr6 | 26674848 | G-T | HOMO | ||
| SBS | Chr6 | 5194423 | A-T | HET | ||
| SBS | Chr6 | 6702612 | A-G | HET | ||
| SBS | Chr7 | 10314299 | G-A | HET | ||
| SBS | Chr7 | 1064925 | C-T | HET | ||
| SBS | Chr7 | 12625680 | G-A | HET | ||
| SBS | Chr7 | 16615279 | T-C | HET | ||
| SBS | Chr7 | 4687855 | G-T | HET | ||
| SBS | Chr8 | 13300224 | C-A | HET | ||
| SBS | Chr8 | 21260287 | G-A | HET | LOC_Os08g33950 | |
| SBS | Chr8 | 2476135 | T-C | HET | ||
| SBS | Chr8 | 7333694 | G-A | HET | ||
| SBS | Chr9 | 13204475 | C-T | HET | ||
| SBS | Chr9 | 13267087 | A-T | HET |
Deletions: 37
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr3 | 185398 | 185398 | 1 | |
| Deletion | Chr10 | 768788 | 768788 | 1 | |
| Deletion | Chr6 | 1571398 | 1571406 | 9 | |
| Deletion | Chr4 | 2987307 | 2987322 | 16 | |
| Deletion | Chr5 | 4201001 | 4546000 | 345000 | 52 |
| Deletion | Chr5 | 4507891 | 4507891 | 1 | |
| Deletion | Chr9 | 4513398 | 4513398 | 1 | LOC_Os09g08630 |
| Deletion | Chr8 | 6526571 | 6526574 | 4 | |
| Deletion | Chr11 | 7596001 | 7682000 | 86000 | 11 |
| Deletion | Chr11 | 7692001 | 7844000 | 152000 | 19 |
| Deletion | Chr11 | 7849001 | 8414000 | 565000 | 83 |
| Deletion | Chr11 | 8415001 | 8428000 | 13000 | 2 |
| Deletion | Chr11 | 8435001 | 8511000 | 76000 | 15 |
| Deletion | Chr5 | 9704337 | 9704337 | 1 | |
| Deletion | Chr5 | 9894684 | 9894684 | 1 | |
| Deletion | Chr8 | 10350542 | 10350542 | 1 | |
| Deletion | Chr8 | 10382955 | 10382955 | 1 | |
| Deletion | Chr9 | 12954529 | 12954532 | 4 | |
| Deletion | Chr5 | 13098204 | 13098210 | 7 | |
| Deletion | Chr9 | 14313214 | 14313214 | 1 | |
| Deletion | Chr2 | 15864435 | 15864435 | 1 | |
| Deletion | Chr1 | 16367345 | 16367346 | 2 | |
| Deletion | Chr2 | 17938541 | 17938541 | 1 | |
| Deletion | Chr5 | 20844486 | 20844486 | 1 | |
| Deletion | Chr10 | 20975571 | 20975574 | 4 | |
| Deletion | Chr5 | 21096927 | 21096928 | 2 | |
| Deletion | Chr9 | 22681727 | 22681727 | 1 | |
| Deletion | Chr5 | 22743065 | 22743074 | 10 | |
| Deletion | Chr4 | 23222958 | 23222958 | 1 | |
| Deletion | Chr1 | 24736563 | 24736565 | 3 | |
| Deletion | Chr1 | 25136150 | 25136154 | 5 | LOC_Os01g43870 |
| Deletion | Chr2 | 25530970 | 25530974 | 5 | |
| Deletion | Chr7 | 28050984 | 28050991 | 8 | |
| Deletion | Chr1 | 28376017 | 28376020 | 4 | |
| Deletion | Chr7 | 29696789 | 29696832 | 44 | |
| Deletion | Chr2 | 30935224 | 30935224 | 1 | |
| Deletion | Chr2 | 32216566 | 32216570 | 5 |
Insertions: 13
Inversions: 2
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr12 | 6600728 | 7334164 | LOC_Os12g13170 |
| Inversion | Chr11 | 8712627 | 9640238 |
Translocations: 3
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr12 | 7333622 | Chr7 | 22722333 | LOC_Os12g13170 |
| Translocation | Chr12 | 7334156 | Chr7 | 22722338 | LOC_Os12g13170 |
| Translocation | Chr11 | 8548490 | Chr3 | 2904210 | LOC_Os11g15170 |
