Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN4769-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN4769-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 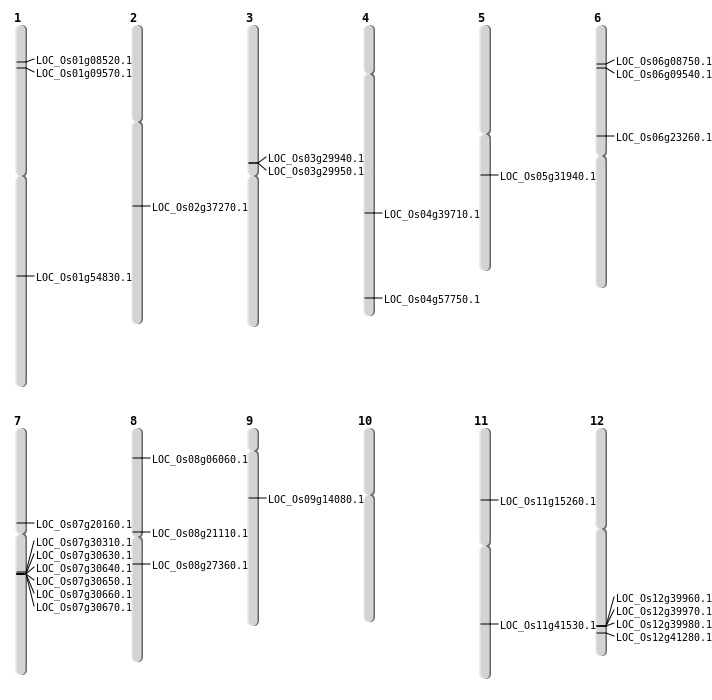
|
| Variant Information |
|---|
Single base substitutions: 72
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 23656462 | A-T | HET | ||
| SBS | Chr1 | 2850991 | G-A | HOMO | ||
| SBS | Chr1 | 31535788 | T-G | HOMO | LOC_Os01g54830 | |
| SBS | Chr1 | 33985559 | G-T | HET | ||
| SBS | Chr1 | 33985560 | C-T | HET | ||
| SBS | Chr1 | 37749790 | G-T | HET | ||
| SBS | Chr1 | 42329271 | T-A | HET | ||
| SBS | Chr10 | 18755173 | A-T | HET | ||
| SBS | Chr10 | 19692534 | G-A | HET | ||
| SBS | Chr10 | 2684884 | C-G | HOMO | ||
| SBS | Chr11 | 10597464 | C-T | HET | ||
| SBS | Chr11 | 18559100 | G-T | HET | ||
| SBS | Chr11 | 24907956 | G-A | HET | LOC_Os11g41530 | |
| SBS | Chr11 | 4120654 | A-T | HET | ||
| SBS | Chr12 | 18892097 | T-C | HOMO | ||
| SBS | Chr12 | 26242539 | C-A | HET | ||
| SBS | Chr12 | 5216396 | G-T | HET | ||
| SBS | Chr2 | 11133387 | C-T | HET | ||
| SBS | Chr2 | 22541250 | C-T | HET | LOC_Os02g37270 | |
| SBS | Chr2 | 22541253 | C-A | HET | LOC_Os02g37270 | |
| SBS | Chr2 | 30840332 | G-A | HET | ||
| SBS | Chr2 | 9979356 | A-G | HET | ||
| SBS | Chr3 | 13453571 | C-T | HET | ||
| SBS | Chr3 | 21943308 | G-A | HET | ||
| SBS | Chr3 | 24465542 | C-T | HET | ||
| SBS | Chr3 | 34121922 | T-C | HET | ||
| SBS | Chr3 | 35913864 | C-G | HET | ||
| SBS | Chr3 | 4245161 | C-T | HET | ||
| SBS | Chr3 | 8406048 | A-T | HET | ||
| SBS | Chr3 | 8853216 | G-C | HET | ||
| SBS | Chr3 | 9513718 | G-T | HET | ||
| SBS | Chr4 | 12846419 | C-A | HET | ||
| SBS | Chr4 | 14014925 | G-T | HET | ||
| SBS | Chr4 | 15086318 | G-C | HET | ||
| SBS | Chr4 | 23103850 | C-A | HET | ||
| SBS | Chr4 | 25273911 | A-T | HET | ||
| SBS | Chr4 | 27014776 | G-C | HET | ||
| SBS | Chr4 | 33119528 | A-G | HOMO | ||
| SBS | Chr4 | 33469095 | T-A | HOMO | ||
| SBS | Chr4 | 6009831 | A-T | HET | ||
| SBS | Chr5 | 14730182 | C-A | HET | ||
| SBS | Chr5 | 17614737 | T-C | HET | ||
| SBS | Chr5 | 18309033 | T-C | HET | ||
| SBS | Chr5 | 23888972 | A-C | HET | ||
| SBS | Chr5 | 25880448 | G-T | HET | ||
| SBS | Chr5 | 26607133 | C-T | HET | ||
| SBS | Chr5 | 8644403 | G-T | HET | ||
| SBS | Chr5 | 8644404 | C-T | HET | ||
| SBS | Chr6 | 28614718 | G-T | HET | ||
| SBS | Chr6 | 28614719 | A-C | HET | ||
| SBS | Chr6 | 30517096 | C-T | HOMO | ||
| SBS | Chr6 | 5472367 | C-T | HET | ||
| SBS | Chr6 | 5748404 | C-T | HOMO | ||
| SBS | Chr6 | 9538736 | G-C | HET | ||
| SBS | Chr7 | 17918494 | G-A | HET | LOC_Os07g30310 | |
| SBS | Chr7 | 658564 | T-G | HOMO | ||
| SBS | Chr8 | 12613846 | T-A | HOMO | LOC_Os08g21110 | |
| SBS | Chr8 | 16709697 | G-A | HET | LOC_Os08g27360 | |
| SBS | Chr8 | 19631973 | A-G | HET | ||
| SBS | Chr8 | 22050122 | G-A | HET | ||
| SBS | Chr8 | 22525585 | C-G | HET | ||
| SBS | Chr8 | 4923119 | C-T | HET | ||
| SBS | Chr8 | 9976308 | C-T | HET | ||
| SBS | Chr9 | 10250276 | A-T | HET | ||
| SBS | Chr9 | 13520400 | G-A | HET | ||
| SBS | Chr9 | 17006297 | T-A | HET | ||
| SBS | Chr9 | 4237917 | C-T | HOMO | ||
| SBS | Chr9 | 4308627 | T-G | HET | ||
| SBS | Chr9 | 4308633 | A-G | HET | ||
| SBS | Chr9 | 7497508 | C-T | HET | ||
| SBS | Chr9 | 8334567 | C-T | HET | LOC_Os09g14080 | |
| SBS | Chr9 | 9353758 | T-A | HET |
Deletions: 28
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr9 | 92058 | 92058 | 1 | |
| Deletion | Chr7 | 314092 | 314093 | 2 | |
| Deletion | Chr3 | 4013335 | 4013335 | 1 | |
| Deletion | Chr1 | 5761289 | 5761289 | 1 | |
| Deletion | Chr9 | 6887630 | 6887635 | 6 | |
| Deletion | Chr6 | 7084528 | 7084529 | 2 | |
| Deletion | Chr9 | 7125885 | 7125885 | 1 | |
| Deletion | Chr11 | 11112871 | 11112876 | 6 | |
| Deletion | Chr1 | 11195203 | 11195262 | 60 | |
| Deletion | Chr7 | 12229807 | 12229807 | 1 | |
| Deletion | Chr2 | 12332722 | 12332722 | 1 | |
| Deletion | Chr2 | 12387445 | 12387445 | 1 | |
| Deletion | Chr3 | 13432600 | 13432603 | 4 | |
| Deletion | Chr12 | 13440777 | 13440777 | 1 | |
| Deletion | Chr6 | 13580857 | 13580861 | 5 | LOC_Os06g23260 |
| Deletion | Chr8 | 16649140 | 16649157 | 18 | |
| Deletion | Chr1 | 16923019 | 16923184 | 166 | |
| Deletion | Chr9 | 17779354 | 17779354 | 1 | |
| Deletion | Chr7 | 18129001 | 18137000 | 8000 | 3 |
| Deletion | Chr7 | 18141001 | 18155000 | 14000 | 2 |
| Deletion | Chr1 | 18277015 | 18277038 | 24 | |
| Deletion | Chr4 | 23667291 | 23667291 | 1 | LOC_Os04g39710 |
| Deletion | Chr2 | 24386060 | 24386123 | 64 | |
| Deletion | Chr12 | 24694001 | 24709000 | 15000 | 3 |
| Deletion | Chr7 | 27748554 | 27748554 | 1 | |
| Deletion | Chr2 | 28799582 | 28799600 | 19 | |
| Deletion | Chr3 | 34122819 | 34122819 | 1 | |
| Deletion | Chr1 | 35184572 | 35184589 | 18 |
Insertions: 18
Inversions: 4
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr1 | 4215128 | 4907356 | 2 |
| Inversion | Chr1 | 4215134 | 4907364 | 2 |
| Inversion | Chr6 | 4382549 | 4850223 | 2 |
| Inversion | Chr1 | 39486153 | 39486286 |
Translocations: 12
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr6 | 4382536 | Chr3 | 17062480 | 2 |
| Translocation | Chr6 | 4850222 | Chr3 | 17068335 | 2 |
| Translocation | Chr8 | 5822110 | Chr5 | 18605710 | LOC_Os05g31940 |
| Translocation | Chr8 | 5836421 | Chr5 | 18605744 | LOC_Os05g31940 |
| Translocation | Chr9 | 6788427 | Chr3 | 5133173 | |
| Translocation | Chr9 | 7748563 | Chr3 | 5134246 | |
| Translocation | Chr11 | 10852750 | Chr7 | 18129439 | LOC_Os07g30630 |
| Translocation | Chr11 | 10852756 | Chr7 | 18129186 | |
| Translocation | Chr12 | 25600717 | Chr8 | 3310433 | 2 |
| Translocation | Chr12 | 26699113 | Chr8 | 3310421 | LOC_Os08g06060 |
| Translocation | Chr4 | 34397354 | Chr2 | 30120625 | LOC_Os04g57750 |
| Translocation | Chr4 | 34397366 | Chr2 | 30120608 | LOC_Os04g57750 |
