Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN477-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN477-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 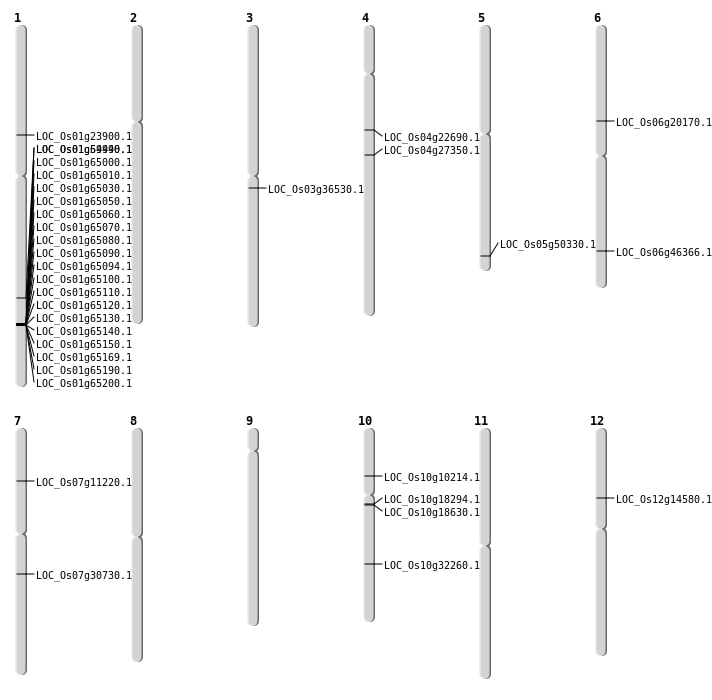
|
| Variant Information |
|---|
Single base substitutions: 30
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 29065522 | C-T | HET | INTERGENIC | |
| SBS | Chr1 | 40163508 | G-A | HET | INTERGENIC | |
| SBS | Chr10 | 13890407 | T-A | HOMO | INTERGENIC | |
| SBS | Chr10 | 1671034 | C-T | HET | SYNONYMOUS_CODING | |
| SBS | Chr10 | 16887120 | C-G | HET | INTERGENIC | |
| SBS | Chr10 | 5619500 | C-T | HOMO | NON_SYNONYMOUS_CODING | LOC_Os10g10214 |
| SBS | Chr10 | 9301158 | T-C | HOMO | NON_SYNONYMOUS_CODING | LOC_Os10g18294 |
| SBS | Chr10 | 9450328 | C-A | HOMO | NON_SYNONYMOUS_CODING | LOC_Os10g18630 |
| SBS | Chr10 | 993871 | C-T | HOMO | INTRON | |
| SBS | Chr11 | 16976292 | C-T | HET | INTERGENIC | |
| SBS | Chr11 | 17308906 | T-A | HET | INTERGENIC | |
| SBS | Chr12 | 14061137 | G-C | HOMO | INTERGENIC | |
| SBS | Chr12 | 21720614 | C-A | HOMO | INTERGENIC | |
| SBS | Chr2 | 32063255 | G-T | HOMO | INTRON | |
| SBS | Chr3 | 14007032 | C-T | HET | SYNONYMOUS_CODING | |
| SBS | Chr3 | 29024780 | T-C | HET | INTERGENIC | |
| SBS | Chr3 | 6703116 | T-C | HET | INTRON | |
| SBS | Chr3 | 6703117 | A-T | HET | INTRON | |
| SBS | Chr3 | 9863238 | T-C | HOMO | INTERGENIC | |
| SBS | Chr4 | 18479586 | T-C | HET | INTERGENIC | |
| SBS | Chr6 | 10972061 | T-A | HET | INTRON | |
| SBS | Chr6 | 11571504 | G-T | HET | NON_SYNONYMOUS_CODING | LOC_Os06g20170 |
| SBS | Chr6 | 15066808 | A-G | HET | INTRON | |
| SBS | Chr6 | 2156930 | T-C | HET | INTERGENIC | |
| SBS | Chr6 | 29080753 | A-T | HET | INTERGENIC | |
| SBS | Chr6 | 8379397 | T-A | HOMO | INTERGENIC | |
| SBS | Chr7 | 1065828 | A-G | HOMO | INTERGENIC | |
| SBS | Chr7 | 6179473 | G-A | HET | STOP_GAINED | LOC_Os07g11220 |
| SBS | Chr8 | 7933572 | C-A | HET | INTERGENIC | |
| SBS | Chr9 | 22604201 | A-T | HET | INTRON |
Deletions: 25
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr12 | 372147 | 372152 | 5 | |
| Deletion | Chr6 | 2306696 | 2306702 | 6 | |
| Deletion | Chr8 | 5430241 | 5430245 | 4 | |
| Deletion | Chr5 | 5586155 | 5586160 | 5 | |
| Deletion | Chr7 | 6778439 | 6778440 | 1 | |
| Deletion | Chr10 | 7191845 | 7191847 | 2 | |
| Deletion | Chr12 | 8327936 | 8327939 | 3 | LOC_Os12g14580 |
| Deletion | Chr8 | 10226806 | 10226821 | 15 | |
| Deletion | Chr6 | 11300462 | 11300473 | 11 | |
| Deletion | Chr4 | 12863556 | 12863578 | 22 | LOC_Os04g22690 |
| Deletion | Chr6 | 13304202 | 13304238 | 36 | |
| Deletion | Chr1 | 13450253 | 13450255 | 2 | LOC_Os01g23900 |
| Deletion | Chr7 | 15991825 | 15991826 | 1 | |
| Deletion | Chr4 | 16167963 | 16167973 | 10 | LOC_Os04g27350 |
| Deletion | Chr7 | 18190278 | 18190280 | 2 | LOC_Os07g30730 |
| Deletion | Chr3 | 20243492 | 20243494 | 2 | LOC_Os03g36530 |
| Deletion | Chr6 | 20458847 | 20458853 | 6 | |
| Deletion | Chr4 | 22542697 | 22542698 | 1 | |
| Deletion | Chr2 | 24700043 | 24700046 | 3 | |
| Deletion | Chr7 | 25737134 | 25737141 | 7 | |
| Deletion | Chr5 | 28839031 | 28839037 | 6 | LOC_Os05g50330 |
| Deletion | Chr5 | 29749208 | 29749217 | 9 | |
| Deletion | Chr1 | 31267810 | 31267824 | 14 | |
| Deletion | Chr1 | 34381430 | 34381437 | 7 | LOC_Os01g59440 |
| Deletion | Chr1 | 37724001 | 37835000 | 111000 | 19 |
Insertions: 7
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr1 | 21109752 | 21109752 | 1 | |
| Insertion | Chr2 | 8864469 | 8864472 | 4 | |
| Insertion | Chr3 | 12114390 | 12114391 | 2 | |
| Insertion | Chr5 | 28034317 | 28034317 | 1 | |
| Insertion | Chr6 | 28133805 | 28133806 | 2 | LOC_Os06g46366 |
| Insertion | Chr6 | 3999411 | 3999412 | 2 | |
| Insertion | Chr7 | 12674433 | 12674434 | 2 |
Inversions: 1
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr10 | 16935183 | 17381268 | LOC_Os10g32260 |
No Translocation
