Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN482-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN482-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 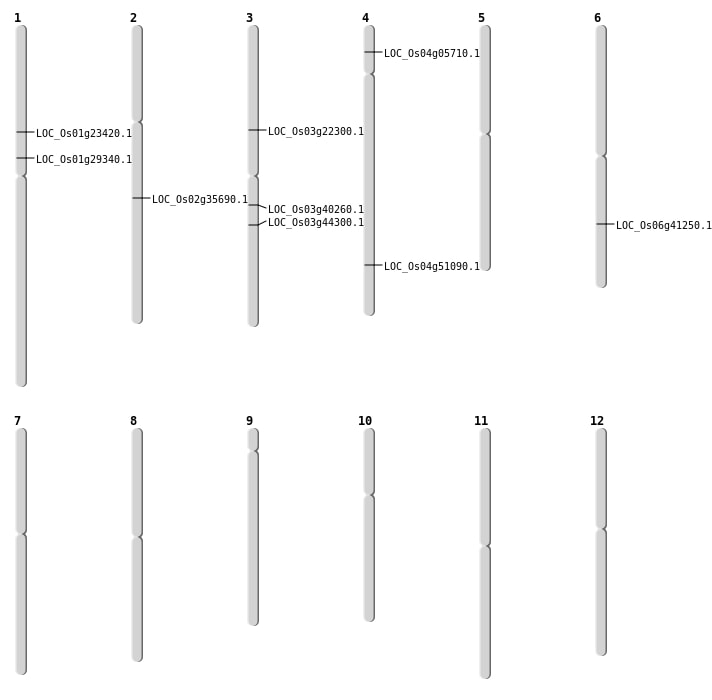
|
| Variant Information |
|---|
Single base substitutions: 30
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 10633372 | G-C | HET | INTERGENIC | |
| SBS | Chr1 | 16440514 | T-C | HET | SYNONYMOUS_CODING | |
| SBS | Chr1 | 23143617 | G-A | HET | INTERGENIC | |
| SBS | Chr1 | 29907667 | T-C | HOMO | INTERGENIC | |
| SBS | Chr1 | 6567405 | A-T | HET | INTRON | |
| SBS | Chr10 | 21843999 | C-A | HET | INTERGENIC | |
| SBS | Chr11 | 10378174 | G-A | HOMO | INTERGENIC | |
| SBS | Chr11 | 15434494 | T-C | HET | INTERGENIC | |
| SBS | Chr12 | 14103085 | G-A | HET | INTERGENIC | |
| SBS | Chr12 | 17453188 | G-T | HET | INTERGENIC | |
| SBS | Chr12 | 24907184 | C-T | HOMO | SYNONYMOUS_CODING | |
| SBS | Chr2 | 16956566 | G-T | HET | INTERGENIC | |
| SBS | Chr2 | 18458837 | C-T | HET | INTERGENIC | |
| SBS | Chr2 | 21451208 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os02g35690 |
| SBS | Chr3 | 12783890 | C-T | HOMO | NON_SYNONYMOUS_CODING | LOC_Os03g22300 |
| SBS | Chr3 | 25180780 | C-T | HOMO | INTERGENIC | |
| SBS | Chr3 | 25218515 | A-T | HOMO | INTERGENIC | |
| SBS | Chr4 | 12228513 | A-G | HET | INTRON | |
| SBS | Chr4 | 2910490 | C-T | HOMO | INTERGENIC | |
| SBS | Chr4 | 2916671 | C-A | HOMO | NON_SYNONYMOUS_CODING | LOC_Os04g05710 |
| SBS | Chr4 | 7599637 | T-A | HET | INTERGENIC | |
| SBS | Chr5 | 2341022 | G-T | HET | INTERGENIC | |
| SBS | Chr6 | 15567300 | T-A | HET | INTERGENIC | |
| SBS | Chr6 | 21873242 | C-T | HET | INTERGENIC | |
| SBS | Chr6 | 27304442 | G-A | HET | INTERGENIC | |
| SBS | Chr6 | 27304443 | T-A | HET | INTERGENIC | |
| SBS | Chr7 | 20607633 | T-G | HET | INTERGENIC | |
| SBS | Chr7 | 22732858 | A-C | HET | INTERGENIC | |
| SBS | Chr8 | 17598255 | G-A | HET | SYNONYMOUS_CODING | |
| SBS | Chr8 | 28247521 | G-A | HOMO | INTERGENIC |
Deletions: 22
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr10 | 517451 | 517458 | 7 | |
| Deletion | Chr4 | 2157114 | 2157129 | 15 | |
| Deletion | Chr1 | 6536535 | 6536538 | 3 | |
| Deletion | Chr2 | 8724960 | 8724969 | 9 | |
| Deletion | Chr11 | 9823599 | 9823610 | 11 | |
| Deletion | Chr1 | 12238188 | 12238190 | 2 | |
| Deletion | Chr1 | 13159313 | 13159319 | 6 | LOC_Os01g23420 |
| Deletion | Chr3 | 15687342 | 15687343 | 1 | |
| Deletion | Chr1 | 16440519 | 16440531 | 12 | LOC_Os01g29340 |
| Deletion | Chr4 | 18884989 | 18884990 | 1 | |
| Deletion | Chr8 | 20985968 | 20985976 | 8 | |
| Deletion | Chr2 | 21308694 | 21308695 | 1 | |
| Deletion | Chr3 | 22379485 | 22379501 | 16 | LOC_Os03g40260 |
| Deletion | Chr1 | 25381841 | 25381842 | 1 | |
| Deletion | Chr2 | 25617059 | 25617060 | 1 | |
| Deletion | Chr4 | 25730017 | 25730034 | 17 | |
| Deletion | Chr4 | 25890972 | 25890994 | 22 | |
| Deletion | Chr3 | 26944585 | 26944595 | 10 | |
| Deletion | Chr4 | 28036994 | 28036999 | 5 | |
| Deletion | Chr3 | 28779173 | 28779189 | 16 | |
| Deletion | Chr4 | 30257572 | 30257577 | 5 | LOC_Os04g51090 |
| Deletion | Chr4 | 32930354 | 32930355 | 1 |
Insertions: 5
Inversions: 2
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr3 | 3797349 | 24903616 | LOC_Os03g44300 |
| Inversion | Chr6 | 24686099 | 25609540 | LOC_Os06g41250 |
Translocations: 4
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr6 | 3234375 | Chr1 | 23346976 | |
| Translocation | Chr11 | 22938433 | Chr6 | 1671809 | |
| Translocation | Chr8 | 27052627 | Chr3 | 3797335 | LOC_Os08g42740 |
| Translocation | Chr8 | 27052639 | Chr3 | 24916396 | LOC_Os08g42740 |
