Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN483-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN483-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 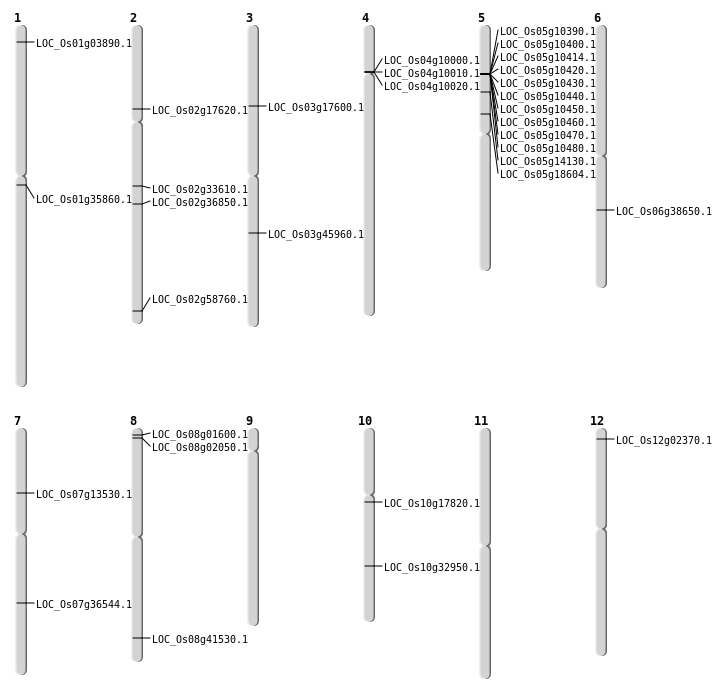
|
| Variant Information |
|---|
Single base substitutions: 42
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 16444546 | C-T | HET | INTERGENIC | |
| SBS | Chr1 | 27964519 | C-T | HET | INTRON | |
| SBS | Chr1 | 28363305 | G-A | HET | INTERGENIC | |
| SBS | Chr1 | 30459127 | G-A | HOMO | INTERGENIC | |
| SBS | Chr1 | 42649473 | C-T | HET | INTRON | |
| SBS | Chr10 | 10354841 | A-G | HOMO | INTERGENIC | |
| SBS | Chr10 | 144661 | T-A | HET | INTERGENIC | |
| SBS | Chr10 | 19751462 | A-C | HET | INTERGENIC | |
| SBS | Chr10 | 4743175 | C-T | HOMO | INTERGENIC | |
| SBS | Chr10 | 7575207 | A-T | HOMO | INTRON | |
| SBS | Chr10 | 8690121 | C-T | HET | INTERGENIC | |
| SBS | Chr10 | 9017351 | G-T | HET | NON_SYNONYMOUS_CODING | LOC_Os10g17820 |
| SBS | Chr12 | 23138528 | A-C | HET | INTERGENIC | |
| SBS | Chr12 | 26839060 | A-T | HOMO | INTERGENIC | |
| SBS | Chr12 | 767351 | C-A | HET | NON_SYNONYMOUS_CODING | LOC_Os12g02370 |
| SBS | Chr2 | 16513223 | C-G | HET | INTERGENIC | |
| SBS | Chr2 | 2647442 | T-G | HET | INTERGENIC | |
| SBS | Chr2 | 35897320 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os02g58760 |
| SBS | Chr3 | 10846547 | A-T | HET | SYNONYMOUS_CODING | |
| SBS | Chr3 | 8588400 | A-T | HET | INTRON | |
| SBS | Chr4 | 193677 | A-T | HET | INTRON | |
| SBS | Chr4 | 23297364 | A-T | HOMO | INTRON | |
| SBS | Chr4 | 25617926 | C-G | HET | INTERGENIC | |
| SBS | Chr4 | 27036883 | C-A | HET | INTRON | |
| SBS | Chr4 | 5376548 | C-T | HET | INTERGENIC | |
| SBS | Chr5 | 18586593 | G-A | HOMO | INTERGENIC | |
| SBS | Chr5 | 5840305 | G-A | HET | INTERGENIC | |
| SBS | Chr5 | 5840306 | G-A | HET | INTERGENIC | |
| SBS | Chr5 | 7893962 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os05g14130 |
| SBS | Chr5 | 8703602 | T-A | HET | INTRON | |
| SBS | Chr5 | 8703609 | G-A | HET | INTRON | |
| SBS | Chr7 | 17779697 | T-A | HET | UTR_3_PRIME | |
| SBS | Chr7 | 23979217 | A-G | HOMO | INTERGENIC | |
| SBS | Chr7 | 28751856 | T-G | HET | INTERGENIC | |
| SBS | Chr8 | 16562924 | C-A | HOMO | INTERGENIC | |
| SBS | Chr8 | 26125408 | T-C | HET | INTERGENIC | |
| SBS | Chr8 | 27560556 | T-G | HET | INTERGENIC | |
| SBS | Chr8 | 8226718 | T-C | HET | INTERGENIC | |
| SBS | Chr8 | 8832103 | G-T | HET | INTERGENIC | |
| SBS | Chr9 | 22762947 | C-A | HET | UTR_5_PRIME | |
| SBS | Chr9 | 3025246 | A-G | HET | INTERGENIC | |
| SBS | Chr9 | 6735634 | G-C | HOMO | INTERGENIC |
Deletions: 26
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr4 | 351665 | 351667 | 2 | |
| Deletion | Chr4 | 361261 | 361270 | 9 | |
| Deletion | Chr1 | 1657970 | 1657978 | 8 | LOC_Os01g03890 |
| Deletion | Chr6 | 4050534 | 4050539 | 5 | |
| Deletion | Chr5 | 4249643 | 4249646 | 3 | |
| Deletion | Chr4 | 5381001 | 5389000 | 8000 | 3 |
| Deletion | Chr5 | 5664001 | 5708000 | 44000 | 10 |
| Deletion | Chr3 | 7450291 | 7450292 | 1 | |
| Deletion | Chr6 | 7496830 | 7496857 | 27 | |
| Deletion | Chr7 | 7744585 | 7744588 | 3 | LOC_Os07g13530 |
| Deletion | Chr5 | 8704347 | 8704753 | 406 | |
| Deletion | Chr5 | 9832590 | 9832591 | 1 | |
| Deletion | Chr5 | 10776438 | 10777125 | 687 | LOC_Os05g18604 |
| Deletion | Chr6 | 12282451 | 12282458 | 7 | |
| Deletion | Chr3 | 16638484 | 16638487 | 3 | |
| Deletion | Chr10 | 17248360 | 17248361 | 1 | LOC_Os10g32950 |
| Deletion | Chr9 | 18391754 | 18391777 | 23 | |
| Deletion | Chr10 | 19088936 | 19089031 | 95 | |
| Deletion | Chr8 | 19173473 | 19173475 | 2 | |
| Deletion | Chr1 | 19857218 | 19857226 | 8 | LOC_Os01g35860 |
| Deletion | Chr10 | 19893417 | 19893425 | 8 | |
| Deletion | Chr2 | 23883658 | 23883659 | 1 | |
| Deletion | Chr3 | 25972291 | 25972316 | 25 | LOC_Os03g45960 |
| Deletion | Chr4 | 27542256 | 27542262 | 6 | |
| Deletion | Chr7 | 28606745 | 28606764 | 19 | |
| Deletion | Chr2 | 34833793 | 34833805 | 12 |
Insertions: 5
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr11 | 6203339 | 6203342 | 4 | |
| Insertion | Chr2 | 10135304 | 10135307 | 4 | LOC_Os02g17620 |
| Insertion | Chr2 | 21026487 | 21026487 | 1 | |
| Insertion | Chr3 | 9791517 | 9791517 | 1 | LOC_Os03g17600 |
| Insertion | Chr6 | 22952191 | 22952195 | 5 | LOC_Os06g38650 |
Inversions: 8
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr8 | 360285 | 664098 | 2 |
| Inversion | Chr7 | 5448421 | 6586515 | |
| Inversion | Chr2 | 18694627 | 21538653 | |
| Inversion | Chr2 | 18695506 | 21538653 | |
| Inversion | Chr2 | 19588627 | 19698134 | |
| Inversion | Chr2 | 19588642 | 19698183 | |
| Inversion | Chr2 | 20013270 | 22235426 | 2 |
| Inversion | Chr7 | 21407753 | 21849461 | LOC_Os07g36544 |
Translocations: 5
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr10 | 1222901 | Chr8 | 16258229 | |
| Translocation | Chr5 | 10776448 | Chr2 | 20013271 | 2 |
| Translocation | Chr5 | 10777108 | Chr2 | 22235415 | 2 |
| Translocation | Chr8 | 26236742 | Chr7 | 5448972 | LOC_Os08g41530 |
| Translocation | Chr8 | 26236743 | Chr7 | 5448431 | LOC_Os08g41530 |
