Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN4853-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN4853-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 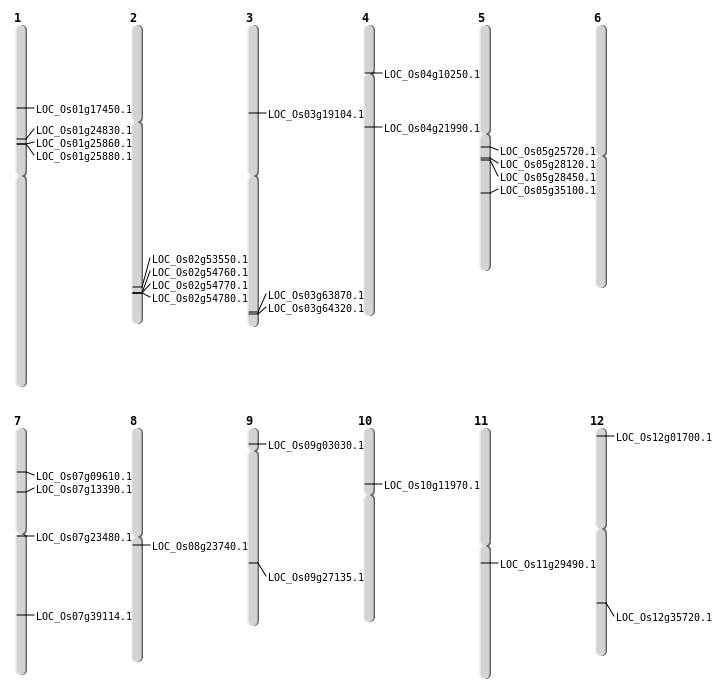
|
| Variant Information |
|---|
Single base substitutions: 50
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 10035219 | A-T | HET | LOC_Os01g17450 | |
| SBS | Chr1 | 13731639 | C-T | HET | ||
| SBS | Chr1 | 13985196 | C-T | HET | LOC_Os01g24830 | |
| SBS | Chr10 | 13933042 | C-T | HOMO | ||
| SBS | Chr10 | 14000611 | T-C | HOMO | ||
| SBS | Chr10 | 14035180 | C-T | HOMO | ||
| SBS | Chr10 | 19263249 | A-G | HET | ||
| SBS | Chr10 | 6635710 | C-T | HET | LOC_Os10g11970 | |
| SBS | Chr11 | 19933453 | A-C | HET | ||
| SBS | Chr11 | 23620610 | C-T | HET | ||
| SBS | Chr12 | 19334884 | C-T | HET | ||
| SBS | Chr12 | 19334890 | A-T | HET | ||
| SBS | Chr12 | 21739452 | A-G | HOMO | LOC_Os12g35720 | |
| SBS | Chr12 | 22703257 | A-T | HET | ||
| SBS | Chr2 | 21125319 | C-T | HET | ||
| SBS | Chr2 | 26244136 | A-T | HET | ||
| SBS | Chr2 | 32765766 | G-A | HET | LOC_Os02g53550 | |
| SBS | Chr2 | 33778219 | G-A | HET | ||
| SBS | Chr3 | 12381002 | G-C | HOMO | ||
| SBS | Chr3 | 22486454 | C-T | HET | ||
| SBS | Chr3 | 31183754 | A-C | HET | ||
| SBS | Chr3 | 32617587 | T-C | HET | ||
| SBS | Chr3 | 35863057 | T-A | HOMO | ||
| SBS | Chr3 | 36351882 | A-G | HET | LOC_Os03g64320 | |
| SBS | Chr3 | 6506541 | G-A | HOMO | ||
| SBS | Chr4 | 12449122 | A-G | HOMO | LOC_Os04g21990 | |
| SBS | Chr4 | 14865278 | A-G | HOMO | ||
| SBS | Chr4 | 17263378 | C-T | HET | ||
| SBS | Chr4 | 5537050 | G-A | HOMO | LOC_Os04g10250 | |
| SBS | Chr4 | 5878626 | T-C | HOMO | ||
| SBS | Chr5 | 14962770 | C-A | HOMO | LOC_Os05g25720 | |
| SBS | Chr5 | 16522586 | C-T | HOMO | ||
| SBS | Chr5 | 18016039 | G-T | HET | ||
| SBS | Chr5 | 19804923 | G-T | HET | ||
| SBS | Chr5 | 20966354 | T-C | HET | ||
| SBS | Chr5 | 8108832 | C-A | HOMO | ||
| SBS | Chr6 | 23859018 | C-A | HET | ||
| SBS | Chr6 | 27820001 | T-A | HET | ||
| SBS | Chr6 | 28034467 | C-T | HET | ||
| SBS | Chr6 | 28761672 | C-T | HET | ||
| SBS | Chr7 | 20903520 | A-C | HET | ||
| SBS | Chr7 | 25370863 | C-T | HET | ||
| SBS | Chr7 | 7680170 | G-A | HET | LOC_Os07g13390 | |
| SBS | Chr8 | 14366299 | G-A | HET | LOC_Os08g23740 | |
| SBS | Chr8 | 19227725 | T-C | HOMO | ||
| SBS | Chr8 | 4514744 | T-A | HET | ||
| SBS | Chr9 | 1424437 | C-T | HET | LOC_Os09g03030 | |
| SBS | Chr9 | 16501551 | C-G | HET | LOC_Os09g27135 | |
| SBS | Chr9 | 22496004 | C-T | HET | ||
| SBS | Chr9 | 5252859 | A-G | HET |
Deletions: 37
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr9 | 324874 | 324874 | 1 | |
| Deletion | Chr12 | 2621558 | 2621560 | 3 | |
| Deletion | Chr2 | 5796875 | 5796967 | 93 | |
| Deletion | Chr8 | 6147675 | 6147677 | 3 | |
| Deletion | Chr4 | 9190294 | 9190294 | 1 | |
| Deletion | Chr10 | 9961797 | 9961801 | 5 | |
| Deletion | Chr3 | 10704635 | 10704642 | 8 | LOC_Os03g19104 |
| Deletion | Chr1 | 11007402 | 11007402 | 1 | |
| Deletion | Chr7 | 12218119 | 12218120 | 2 | |
| Deletion | Chr5 | 12522477 | 12522477 | 1 | |
| Deletion | Chr7 | 13263218 | 13263218 | 1 | LOC_Os07g23480 |
| Deletion | Chr7 | 13525080 | 13525108 | 29 | |
| Deletion | Chr1 | 14668001 | 14681000 | 13000 | 2 |
| Deletion | Chr3 | 15057789 | 15057789 | 1 | |
| Deletion | Chr3 | 15339100 | 15339100 | 1 | |
| Deletion | Chr10 | 15353026 | 15353026 | 1 | |
| Deletion | Chr4 | 15370844 | 15370844 | 1 | |
| Deletion | Chr6 | 15471302 | 15471454 | 153 | |
| Deletion | Chr5 | 16452214 | 16452245 | 32 | LOC_Os05g28120 |
| Deletion | Chr11 | 17110629 | 17110640 | 12 | LOC_Os11g29490 |
| Deletion | Chr8 | 18565879 | 18565905 | 27 | |
| Deletion | Chr6 | 18662317 | 18662317 | 1 | |
| Deletion | Chr9 | 19192845 | 19192854 | 10 | |
| Deletion | Chr4 | 20273734 | 20273734 | 1 | |
| Deletion | Chr8 | 20613842 | 20613933 | 92 | |
| Deletion | Chr5 | 20839914 | 20839914 | 1 | LOC_Os05g35100 |
| Deletion | Chr10 | 20904904 | 20904904 | 1 | |
| Deletion | Chr5 | 21031185 | 21031255 | 71 | |
| Deletion | Chr5 | 21545104 | 21545104 | 1 | |
| Deletion | Chr7 | 23436476 | 23436476 | 1 | LOC_Os07g39114 |
| Deletion | Chr6 | 26763350 | 26763350 | 1 | |
| Deletion | Chr1 | 28807960 | 28807968 | 9 | |
| Deletion | Chr11 | 28882552 | 28882554 | 3 | |
| Deletion | Chr1 | 30152503 | 30152503 | 1 | |
| Deletion | Chr2 | 33535001 | 33548000 | 13000 | 3 |
| Deletion | Chr1 | 35425384 | 35425384 | 1 | |
| Deletion | Chr3 | 36095980 | 36095980 | 1 | LOC_Os03g63870 |
Insertions: 22
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr1 | 15932605 | 15932605 | 1 | |
| Insertion | Chr1 | 16915036 | 16915036 | 1 | |
| Insertion | Chr1 | 9996373 | 9996373 | 1 | |
| Insertion | Chr10 | 2799446 | 2799446 | 1 | |
| Insertion | Chr10 | 9989957 | 9989962 | 6 | |
| Insertion | Chr11 | 12094442 | 12094498 | 57 | |
| Insertion | Chr11 | 15380594 | 15380652 | 59 | |
| Insertion | Chr11 | 18977940 | 18978012 | 73 | |
| Insertion | Chr12 | 418947 | 419056 | 110 | LOC_Os12g01700 |
| Insertion | Chr12 | 4691131 | 4691132 | 2 | |
| Insertion | Chr2 | 20067832 | 20067832 | 1 | |
| Insertion | Chr2 | 6767052 | 6767052 | 1 | |
| Insertion | Chr2 | 9359458 | 9359458 | 1 | |
| Insertion | Chr3 | 15394250 | 15394250 | 1 | |
| Insertion | Chr4 | 10895202 | 10895202 | 1 | |
| Insertion | Chr4 | 27189648 | 27189648 | 1 | |
| Insertion | Chr4 | 3940886 | 3940886 | 1 | |
| Insertion | Chr5 | 11965739 | 11965742 | 4 | |
| Insertion | Chr5 | 16384705 | 16384713 | 9 | |
| Insertion | Chr5 | 21261086 | 21261086 | 1 | |
| Insertion | Chr8 | 10015031 | 10015034 | 4 | |
| Insertion | Chr9 | 1023000 | 1023000 | 1 |
No Inversion
Translocations: 2
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr10 | 8825400 | Chr9 | 22024982 | |
| Translocation | Chr10 | 8825409 | Chr9 | 22024251 |
