Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN486-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN486-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 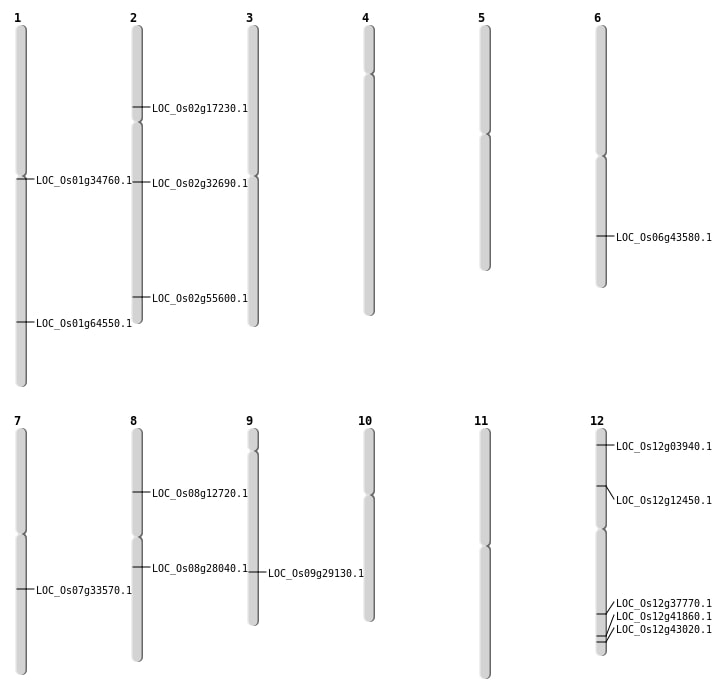
|
| Variant Information |
|---|
Single base substitutions: 31
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 21058758 | C-T | HET | INTERGENIC | |
| SBS | Chr10 | 10778828 | C-A | HOMO | INTERGENIC | |
| SBS | Chr10 | 13291615 | A-T | HET | INTRON | |
| SBS | Chr10 | 2200180 | G-A | HET | INTERGENIC | |
| SBS | Chr11 | 10053709 | C-T | HET | INTERGENIC | |
| SBS | Chr11 | 10372372 | T-A | HET | INTRON | |
| SBS | Chr12 | 17592050 | C-T | HET | INTRON | |
| SBS | Chr2 | 35269024 | A-G | HET | INTERGENIC | |
| SBS | Chr2 | 35723251 | G-A | HET | INTRON | |
| SBS | Chr2 | 8885142 | G-C | HOMO | INTERGENIC | |
| SBS | Chr2 | 9876405 | G-T | HOMO | INTERGENIC | |
| SBS | Chr3 | 8037179 | G-A | HET | INTERGENIC | |
| SBS | Chr3 | 9959620 | C-A | HOMO | INTERGENIC | |
| SBS | Chr4 | 15949885 | G-T | HOMO | SYNONYMOUS_CODING | |
| SBS | Chr5 | 18653857 | A-G | HOMO | INTRON | |
| SBS | Chr5 | 27132970 | A-G | HET | INTRON | |
| SBS | Chr6 | 13308438 | C-T | HET | INTERGENIC | |
| SBS | Chr6 | 13308439 | G-T | HET | INTERGENIC | |
| SBS | Chr6 | 20343615 | C-T | HET | INTERGENIC | |
| SBS | Chr6 | 23800688 | T-A | HET | INTERGENIC | |
| SBS | Chr6 | 26209172 | G-T | HET | SPLICE_SITE_ACCEPTOR | LOC_Os06g43580 |
| SBS | Chr6 | 9538157 | C-A | HET | INTERGENIC | |
| SBS | Chr7 | 20056705 | C-A | HOMO | NON_SYNONYMOUS_CODING | LOC_Os07g33570 |
| SBS | Chr8 | 12303359 | T-C | HOMO | INTERGENIC | |
| SBS | Chr8 | 17081579 | G-C | HOMO | NON_SYNONYMOUS_CODING | LOC_Os08g28040 |
| SBS | Chr8 | 26853943 | C-T | HET | INTERGENIC | |
| SBS | Chr8 | 27943503 | A-T | HET | INTRON | |
| SBS | Chr8 | 7537754 | C-A | HOMO | NON_SYNONYMOUS_CODING | LOC_Os08g12720 |
| SBS | Chr9 | 11466222 | G-C | HET | INTERGENIC | |
| SBS | Chr9 | 17888589 | A-G | HET | INTERGENIC | |
| SBS | Chr9 | 3357329 | C-T | HET | INTERGENIC |
Deletions: 20
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr8 | 333909 | 333910 | 1 | |
| Deletion | Chr12 | 456168 | 456169 | 1 | |
| Deletion | Chr8 | 4102996 | 4103004 | 8 | |
| Deletion | Chr12 | 6850684 | 6850696 | 12 | LOC_Os12g12450 |
| Deletion | Chr2 | 8067005 | 8067030 | 25 | |
| Deletion | Chr2 | 9879007 | 9879216 | 209 | LOC_Os02g17230 |
| Deletion | Chr2 | 10018534 | 10020192 | 1658 | |
| Deletion | Chr2 | 10166278 | 10166284 | 6 | |
| Deletion | Chr10 | 13287145 | 13287146 | 1 | |
| Deletion | Chr11 | 15733665 | 15733668 | 3 | |
| Deletion | Chr2 | 15990245 | 15990247 | 2 | |
| Deletion | Chr12 | 17829927 | 17829929 | 2 | |
| Deletion | Chr12 | 23190755 | 23190756 | 1 | LOC_Os12g37770 |
| Deletion | Chr12 | 23207079 | 23207082 | 3 | |
| Deletion | Chr12 | 26726641 | 26726652 | 11 | LOC_Os12g43020 |
| Deletion | Chr4 | 27274503 | 27274505 | 2 | |
| Deletion | Chr3 | 27992317 | 27992320 | 3 | |
| Deletion | Chr2 | 28311801 | 28311802 | 1 | |
| Deletion | Chr1 | 29335069 | 29335543 | 474 | |
| Deletion | Chr1 | 37455423 | 37455431 | 8 | LOC_Os01g64550 |
Insertions: 5
Inversions: 3
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr9 | 17705608 | 17793895 | LOC_Os09g29130 |
| Inversion | Chr2 | 33970499 | 34053825 | LOC_Os02g55600 |
| Inversion | Chr2 | 33970514 | 34053826 | LOC_Os02g55600 |
Translocations: 5
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr12 | 1631172 | Chr11 | 10418312 | LOC_Os12g03940 |
| Translocation | Chr11 | 6261411 | Chr1 | 29335090 | |
| Translocation | Chr12 | 8582209 | Chr1 | 19169592 | LOC_Os01g34760 |
| Translocation | Chr2 | 19415623 | Chr1 | 7228880 | LOC_Os02g32690 |
| Translocation | Chr12 | 25927137 | Chr2 | 10020236 | 2 |
