Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN494-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN494-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 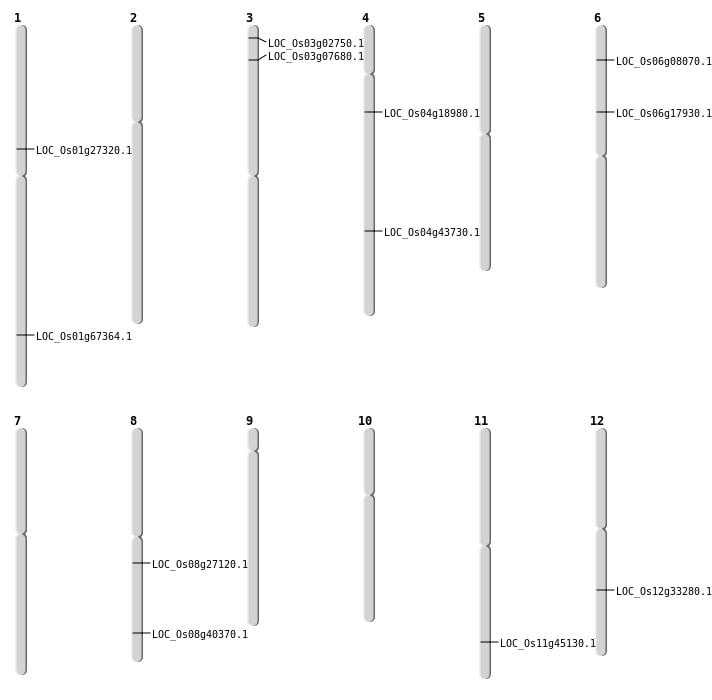
|
| Variant Information |
|---|
Single base substitutions: 34
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 13719910 | T-C | HOMO | INTERGENIC | |
| SBS | Chr1 | 14675904 | T-C | HOMO | INTERGENIC | |
| SBS | Chr1 | 15247579 | G-A | HOMO | NON_SYNONYMOUS_CODING | LOC_Os01g27320 |
| SBS | Chr1 | 15248113 | C-A | HOMO | NON_SYNONYMOUS_CODING | LOC_Os01g27320 |
| SBS | Chr1 | 41385197 | C-T | HOMO | INTRON | |
| SBS | Chr10 | 6720514 | T-A | HET | INTERGENIC | |
| SBS | Chr11 | 10483476 | C-G | HOMO | INTERGENIC | |
| SBS | Chr11 | 27321491 | A-T | HET | NON_SYNONYMOUS_CODING | LOC_Os11g45130 |
| SBS | Chr11 | 9939322 | G-C | HOMO | INTERGENIC | |
| SBS | Chr12 | 15419689 | T-C | HET | INTERGENIC | |
| SBS | Chr12 | 17063681 | C-T | HOMO | INTERGENIC | |
| SBS | Chr2 | 16571054 | C-T | HET | INTERGENIC | |
| SBS | Chr2 | 877767 | C-T | HET | INTRON | |
| SBS | Chr2 | 9597062 | C-T | HET | INTERGENIC | |
| SBS | Chr3 | 1034319 | G-T | HOMO | NON_SYNONYMOUS_CODING | LOC_Os03g02750 |
| SBS | Chr3 | 21582374 | G-C | HET | INTERGENIC | |
| SBS | Chr3 | 34712106 | C-T | HET | INTERGENIC | |
| SBS | Chr4 | 10549261 | T-A | HET | NON_SYNONYMOUS_CODING | LOC_Os04g18980 |
| SBS | Chr5 | 23031233 | G-A | HET | INTRON | |
| SBS | Chr5 | 9371269 | C-T | HET | INTERGENIC | |
| SBS | Chr6 | 11844796 | G-C | HET | INTERGENIC | |
| SBS | Chr6 | 14270898 | C-T | HOMO | INTERGENIC | |
| SBS | Chr6 | 22413038 | T-G | HOMO | INTERGENIC | |
| SBS | Chr6 | 25433218 | G-T | HET | INTERGENIC | |
| SBS | Chr6 | 30786817 | A-T | HET | INTERGENIC | |
| SBS | Chr7 | 26169714 | G-A | HOMO | INTERGENIC | |
| SBS | Chr8 | 10019527 | T-G | HET | INTERGENIC | |
| SBS | Chr8 | 13442788 | G-T | HET | INTERGENIC | |
| SBS | Chr8 | 16573057 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os08g27120 |
| SBS | Chr8 | 25559460 | C-A | HET | NON_SYNONYMOUS_CODING | LOC_Os08g40370 |
| SBS | Chr8 | 3176 | C-T | HOMO | INTERGENIC | |
| SBS | Chr8 | 5376024 | G-T | HET | SYNONYMOUS_CODING | |
| SBS | Chr9 | 19794790 | A-T | HET | INTERGENIC | |
| SBS | Chr9 | 59499 | G-T | HET | INTERGENIC |
Deletions: 24
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr6 | 3907069 | 3907071 | 2 | LOC_Os06g08070 |
| Deletion | Chr7 | 5035497 | 5035512 | 15 | |
| Deletion | Chr4 | 5521845 | 5521847 | 2 | |
| Deletion | Chr5 | 5738129 | 5738159 | 30 | |
| Deletion | Chr6 | 8753492 | 8753501 | 9 | |
| Deletion | Chr6 | 10423667 | 10423983 | 316 | |
| Deletion | Chr3 | 11360859 | 11360865 | 6 | |
| Deletion | Chr4 | 13092623 | 13093135 | 512 | |
| Deletion | Chr10 | 17094620 | 17094627 | 7 | |
| Deletion | Chr8 | 17269880 | 17269882 | 2 | |
| Deletion | Chr7 | 17646569 | 17646595 | 26 | |
| Deletion | Chr12 | 17795208 | 17795215 | 7 | |
| Deletion | Chr9 | 19468296 | 19468297 | 1 | |
| Deletion | Chr6 | 20337816 | 20337829 | 13 | |
| Deletion | Chr7 | 22838278 | 22838279 | 1 | |
| Deletion | Chr7 | 25014618 | 25014625 | 7 | |
| Deletion | Chr11 | 25799782 | 25799789 | 7 | |
| Deletion | Chr4 | 25886459 | 25886489 | 30 | LOC_Os04g43730 |
| Deletion | Chr4 | 25902602 | 25902608 | 6 | |
| Deletion | Chr8 | 26013511 | 26013512 | 1 | |
| Deletion | Chr7 | 27112606 | 27112628 | 22 | |
| Deletion | Chr4 | 30540327 | 30540328 | 1 | |
| Deletion | Chr3 | 34068227 | 34068231 | 4 | |
| Deletion | Chr1 | 35503316 | 35503322 | 6 |
Insertions: 6
Inversions: 3
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr12 | 1914923 | 8868111 | |
| Inversion | Chr12 | 1914948 | 8884886 | |
| Inversion | Chr1 | 39117073 | 39117196 | LOC_Os01g67364 |
Translocations: 7
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr3 | 3902011 | Chr1 | 41304730 | LOC_Os03g07680 |
| Translocation | Chr3 | 3902115 | Chr1 | 41304744 | LOC_Os03g07680 |
| Translocation | Chr6 | 10423675 | Chr2 | 28715310 | LOC_Os06g17930 |
| Translocation | Chr6 | 10423979 | Chr2 | 28715281 | LOC_Os06g17930 |
| Translocation | Chr11 | 13095234 | Chr5 | 20699939 | |
| Translocation | Chr12 | 20138349 | Chr5 | 13277467 | LOC_Os12g33280 |
| Translocation | Chr10 | 22679747 | Chr5 | 16845607 |
