Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN497-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN497-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 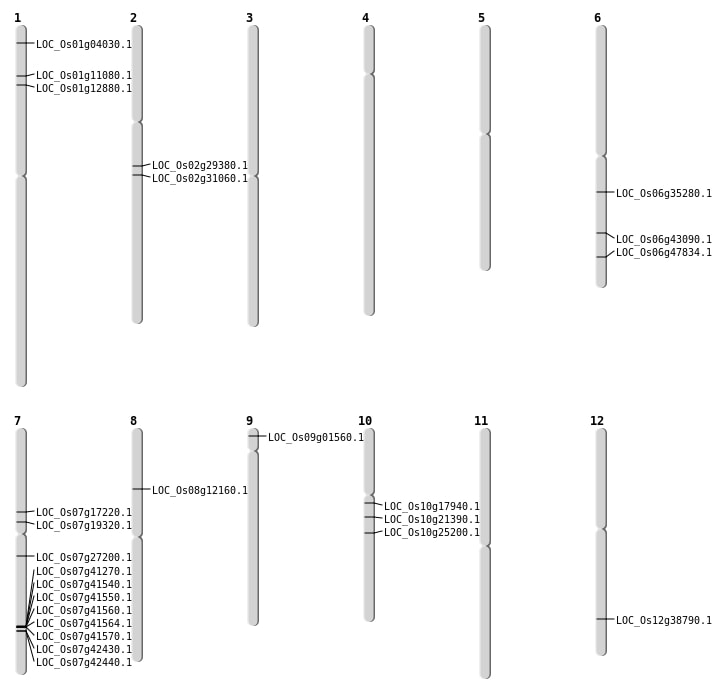
|
| Variant Information |
|---|
Single base substitutions: 46
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 12480024 | C-T | HOMO | INTRON | |
| SBS | Chr1 | 18799251 | T-C | HET | INTERGENIC | |
| SBS | Chr1 | 5918512 | G-A | HOMO | NON_SYNONYMOUS_CODING | LOC_Os01g11080 |
| SBS | Chr1 | 6915066 | A-T | HOMO | UTR_5_PRIME | |
| SBS | Chr10 | 10417002 | T-C | HET | INTRON | |
| SBS | Chr10 | 16342588 | A-G | HET | INTERGENIC | |
| SBS | Chr10 | 22378616 | C-A | HOMO | INTERGENIC | |
| SBS | Chr10 | 4365086 | G-T | HET | INTERGENIC | |
| SBS | Chr10 | 8159500 | A-G | HET | INTERGENIC | |
| SBS | Chr10 | 8311094 | T-A | HET | INTERGENIC | |
| SBS | Chr11 | 20459456 | A-G | HET | INTRON | |
| SBS | Chr11 | 2948516 | G-A | HET | INTERGENIC | |
| SBS | Chr11 | 9207577 | C-T | HET | INTERGENIC | |
| SBS | Chr12 | 23831875 | C-A | HET | NON_SYNONYMOUS_CODING | LOC_Os12g38790 |
| SBS | Chr12 | 8264352 | C-T | HET | INTERGENIC | |
| SBS | Chr2 | 24242129 | T-A | HOMO | INTRON | |
| SBS | Chr3 | 25359798 | C-T | HET | INTERGENIC | |
| SBS | Chr4 | 16490646 | A-C | HET | INTERGENIC | |
| SBS | Chr4 | 16490653 | T-G | HET | INTERGENIC | |
| SBS | Chr4 | 1787471 | C-T | HET | INTERGENIC | |
| SBS | Chr4 | 23338071 | G-T | HOMO | INTERGENIC | |
| SBS | Chr4 | 6879465 | T-C | HET | INTERGENIC | |
| SBS | Chr4 | 9537826 | C-T | HET | SYNONYMOUS_CODING | |
| SBS | Chr5 | 20526044 | G-A | HET | INTERGENIC | |
| SBS | Chr5 | 4668200 | C-T | HET | INTERGENIC | |
| SBS | Chr6 | 10633075 | C-T | HET | INTERGENIC | |
| SBS | Chr6 | 20574792 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os06g35280 |
| SBS | Chr6 | 60259 | A-G | HET | INTERGENIC | |
| SBS | Chr6 | 724767 | A-T | HOMO | INTERGENIC | |
| SBS | Chr7 | 11432670 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os07g19320 |
| SBS | Chr7 | 15142861 | G-A | HET | SYNONYMOUS_CODING | |
| SBS | Chr7 | 2575327 | G-A | HET | INTERGENIC | |
| SBS | Chr7 | 5490354 | C-T | HET | INTERGENIC | |
| SBS | Chr7 | 9211739 | G-A | HET | INTRON | |
| SBS | Chr8 | 1343838 | C-T | HOMO | INTERGENIC | |
| SBS | Chr8 | 18660253 | A-G | HOMO | SYNONYMOUS_CODING | |
| SBS | Chr8 | 24349086 | A-G | HOMO | INTERGENIC | |
| SBS | Chr8 | 25304134 | A-G | HOMO | INTERGENIC | |
| SBS | Chr8 | 2596687 | C-T | HOMO | INTERGENIC | |
| SBS | Chr8 | 5519770 | T-A | HET | INTERGENIC | |
| SBS | Chr8 | 5519771 | A-C | HET | INTERGENIC | |
| SBS | Chr8 | 7461257 | T-A | HET | INTERGENIC | |
| SBS | Chr8 | 7461258 | C-T | HET | INTERGENIC | |
| SBS | Chr9 | 400937 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os09g01560 |
| SBS | Chr9 | 7260586 | T-C | HOMO | INTERGENIC | |
| SBS | Chr9 | 9419816 | C-T | HOMO | INTERGENIC |
Deletions: 19
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr4 | 4567309 | 4567314 | 5 | |
| Deletion | Chr4 | 6484347 | 6484352 | 5 | |
| Deletion | Chr9 | 7901870 | 7901874 | 4 | |
| Deletion | Chr7 | 10142440 | 10142461 | 21 | LOC_Os07g17220 |
| Deletion | Chr10 | 10946193 | 10946195 | 2 | LOC_Os10g21390 |
| Deletion | Chr9 | 12932235 | 12932236 | 1 | |
| Deletion | Chr8 | 14038305 | 14038307 | 2 | |
| Deletion | Chr12 | 14102082 | 14102083 | 1 | |
| Deletion | Chr7 | 15811478 | 15813585 | 2107 | LOC_Os07g27200 |
| Deletion | Chr3 | 18721319 | 18721326 | 7 | |
| Deletion | Chr4 | 23331870 | 23331872 | 2 | |
| Deletion | Chr1 | 23670653 | 23670657 | 4 | |
| Deletion | Chr1 | 23710607 | 23710608 | 1 | |
| Deletion | Chr5 | 23859520 | 23861247 | 1727 | |
| Deletion | Chr7 | 24291785 | 24292512 | 727 | |
| Deletion | Chr7 | 24902500 | 24913176 | 10676 | 5 |
| Deletion | Chr2 | 25479712 | 25479713 | 1 | |
| Deletion | Chr3 | 28923212 | 28923213 | 1 | |
| Deletion | Chr6 | 28952245 | 28953396 | 1151 | LOC_Os06g47834 |
Insertions: 4
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr2 | 18556721 | 18556726 | 6 | LOC_Os02g31060 |
| Insertion | Chr4 | 34449767 | 34449767 | 1 | |
| Insertion | Chr6 | 23312008 | 23312019 | 12 | |
| Insertion | Chr8 | 18779120 | 18779121 | 2 |
Inversions: 6
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr1 | 1756160 | 1793064 | LOC_Os01g04030 |
| Inversion | Chr1 | 6125871 | 7135910 | LOC_Os01g12880 |
| Inversion | Chr10 | 13008638 | 13120884 | LOC_Os10g25200 |
| Inversion | Chr8 | 20489975 | 20761121 | |
| Inversion | Chr7 | 24734230 | 25411185 | 2 |
| Inversion | Chr7 | 24902119 | 25405060 | 2 |
Translocations: 12
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr8 | 7165821 | Chr7 | 25405061 | 2 |
| Translocation | Chr10 | 9079951 | Chr7 | 24902504 | 2 |
| Translocation | Chr10 | 9079958 | Chr7 | 24913204 | 2 |
| Translocation | Chr10 | 13901025 | Chr6 | 28953420 | LOC_Os06g47834 |
| Translocation | Chr10 | 13901031 | Chr6 | 28952249 | LOC_Os06g47834 |
| Translocation | Chr7 | 24291801 | Chr3 | 6038268 | |
| Translocation | Chr7 | 24292107 | Chr3 | 6038265 | |
| Translocation | Chr7 | 24292498 | Chr1 | 4833244 | |
| Translocation | Chr7 | 24902131 | Chr2 | 17449738 | 2 |
| Translocation | Chr7 | 24913925 | Chr2 | 17449734 | 2 |
| Translocation | Chr6 | 25899909 | Chr5 | 23861243 | LOC_Os06g43090 |
| Translocation | Chr6 | 25899935 | Chr5 | 23859528 | LOC_Os06g43090 |
