Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN501-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN501-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 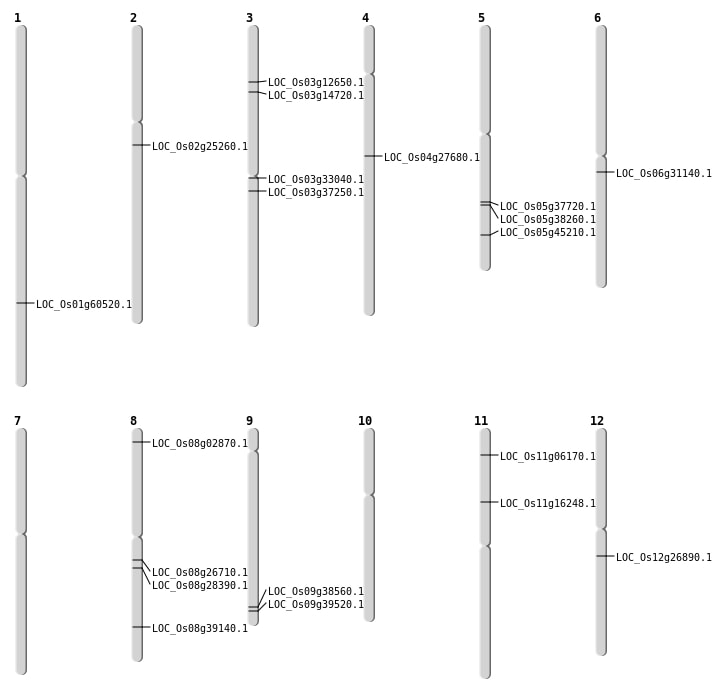
|
| Variant Information |
|---|
Single base substitutions: 33
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 14757078 | T-A | HET | INTRON | |
| SBS | Chr10 | 18380577 | C-T | HOMO | INTERGENIC | |
| SBS | Chr10 | 22362595 | C-T | HOMO | INTERGENIC | |
| SBS | Chr10 | 4490047 | G-C | HOMO | INTERGENIC | |
| SBS | Chr10 | 6667519 | A-G | HET | INTRON | |
| SBS | Chr10 | 7992525 | T-G | HOMO | INTERGENIC | |
| SBS | Chr11 | 21036215 | C-T | HOMO | SYNONYMOUS_CODING | |
| SBS | Chr12 | 13536949 | G-C | HET | INTERGENIC | |
| SBS | Chr2 | 14708285 | G-A | HET | STOP_GAINED | LOC_Os02g25260 |
| SBS | Chr2 | 5552368 | A-G | HET | INTERGENIC | |
| SBS | Chr3 | 14158330 | T-A | HET | INTRON | |
| SBS | Chr3 | 17164073 | A-G | HET | INTRON | |
| SBS | Chr3 | 18896686 | C-T | HOMO | NON_SYNONYMOUS_CODING | LOC_Os03g33040 |
| SBS | Chr3 | 6725096 | G-T | HET | INTERGENIC | |
| SBS | Chr4 | 20777926 | G-T | HOMO | INTERGENIC | |
| SBS | Chr5 | 26240465 | T-G | HET | NON_SYNONYMOUS_CODING | LOC_Os05g45210 |
| SBS | Chr6 | 18092851 | A-C | HET | NON_SYNONYMOUS_CODING | LOC_Os06g31140 |
| SBS | Chr6 | 2435792 | T-C | HET | INTRON | |
| SBS | Chr6 | 2597821 | C-A | HET | INTERGENIC | |
| SBS | Chr7 | 19743004 | G-A | HOMO | INTERGENIC | |
| SBS | Chr7 | 19743007 | G-A | HOMO | INTERGENIC | |
| SBS | Chr7 | 26479962 | T-A | HET | INTERGENIC | |
| SBS | Chr7 | 28002985 | T-C | HOMO | INTRON | |
| SBS | Chr7 | 6129785 | G-T | HET | INTERGENIC | |
| SBS | Chr7 | 6924803 | A-T | HET | INTERGENIC | |
| SBS | Chr8 | 16247298 | T-A | HET | STOP_GAINED | LOC_Os08g26710 |
| SBS | Chr8 | 20671425 | A-G | HET | SPLICE_SITE_REGION | |
| SBS | Chr8 | 6992808 | T-C | HET | INTRON | |
| SBS | Chr9 | 14370885 | T-A | HET | INTRON | |
| SBS | Chr9 | 15488033 | G-A | HET | SYNONYMOUS_CODING | |
| SBS | Chr9 | 19021921 | G-A | HET | INTRON | |
| SBS | Chr9 | 22697818 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os09g39520 |
| SBS | Chr9 | 3444509 | C-T | HET | INTERGENIC |
Deletions: 21
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr12 | 377649 | 377680 | 31 | |
| Deletion | Chr11 | 2939606 | 2939607 | 1 | LOC_Os11g06170 |
| Deletion | Chr6 | 5685874 | 5685884 | 10 | |
| Deletion | Chr3 | 6736449 | 6736455 | 6 | LOC_Os03g12650 |
| Deletion | Chr10 | 7960393 | 7960397 | 4 | |
| Deletion | Chr3 | 8008072 | 8008077 | 5 | LOC_Os03g14720 |
| Deletion | Chr9 | 8976503 | 8976511 | 8 | |
| Deletion | Chr10 | 9613926 | 9613935 | 9 | |
| Deletion | Chr4 | 12954300 | 12954302 | 2 | |
| Deletion | Chr9 | 13821960 | 13821961 | 1 | |
| Deletion | Chr11 | 15514388 | 15514389 | 1 | |
| Deletion | Chr9 | 15718166 | 15718172 | 6 | |
| Deletion | Chr12 | 15764392 | 15764397 | 5 | LOC_Os12g26890 |
| Deletion | Chr3 | 20645883 | 20645891 | 8 | LOC_Os03g37250 |
| Deletion | Chr9 | 22181848 | 22181854 | 6 | LOC_Os09g38560 |
| Deletion | Chr1 | 22638248 | 22638249 | 1 | |
| Deletion | Chr4 | 22905876 | 22905878 | 2 | |
| Deletion | Chr2 | 25989735 | 25989737 | 2 | |
| Deletion | Chr5 | 26316860 | 26316866 | 6 | |
| Deletion | Chr6 | 26952516 | 26952524 | 8 | |
| Deletion | Chr2 | 33985898 | 33985900 | 2 |
Insertions: 3
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr10 | 21922368 | 21922369 | 2 | |
| Insertion | Chr11 | 19639316 | 19639327 | 12 | |
| Insertion | Chr2 | 15646552 | 15646552 | 1 |
Inversions: 8
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr8 | 12100294 | 17314820 | LOC_Os08g28390 |
| Inversion | Chr8 | 17190439 | 17314811 | LOC_Os08g28390 |
| Inversion | Chr8 | 17190453 | 17314821 | LOC_Os08g28390 |
| Inversion | Chr5 | 22079026 | 22660675 | LOC_Os05g37720 |
| Inversion | Chr5 | 22079038 | 22658981 | LOC_Os05g37720 |
| Inversion | Chr5 | 22433512 | 22687714 | LOC_Os05g38260 |
| Inversion | Chr5 | 22658974 | 22687877 | |
| Inversion | Chr1 | 35001387 | 35057865 | LOC_Os01g60520 |
Translocations: 4
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr11 | 8939854 | Chr8 | 16045154 | LOC_Os11g16248 |
| Translocation | Chr4 | 16363770 | Chr1 | 23524841 | LOC_Os04g27680 |
| Translocation | Chr8 | 24720650 | Chr3 | 24395745 | LOC_Os08g39140 |
| Translocation | Chr11 | 26925423 | Chr8 | 1219481 | LOC_Os08g02870 |
