Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN503-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN503-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 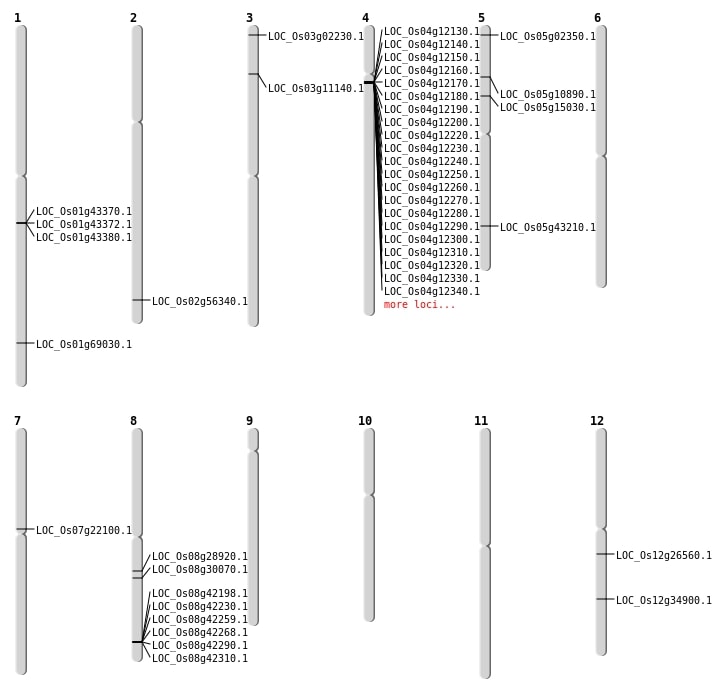
|
| Variant Information |
|---|
Single base substitutions: 32
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 12370114 | C-T | HET | INTERGENIC | |
| SBS | Chr1 | 21469563 | C-T | HET | INTERGENIC | |
| SBS | Chr1 | 26422815 | C-T | HOMO | INTERGENIC | |
| SBS | Chr1 | 3688320 | G-T | HET | INTERGENIC | |
| SBS | Chr1 | 8537912 | T-C | HET | INTERGENIC | |
| SBS | Chr10 | 2303754 | C-T | HOMO | INTERGENIC | |
| SBS | Chr10 | 7833931 | C-T | HET | INTRON | |
| SBS | Chr11 | 20494168 | G-A | HET | INTERGENIC | |
| SBS | Chr11 | 23497659 | C-G | HET | INTERGENIC | |
| SBS | Chr12 | 10991695 | G-A | HET | INTERGENIC | |
| SBS | Chr2 | 25331494 | G-A | HET | INTRON | |
| SBS | Chr2 | 34488797 | T-A | HET | NON_SYNONYMOUS_CODING | LOC_Os02g56340 |
| SBS | Chr3 | 13163622 | C-A | HET | SYNONYMOUS_CODING | |
| SBS | Chr3 | 25981579 | G-A | HOMO | INTRON | |
| SBS | Chr3 | 26280038 | T-C | HET | INTERGENIC | |
| SBS | Chr3 | 28161928 | G-A | HOMO | INTERGENIC | |
| SBS | Chr4 | 3439630 | C-A | HOMO | INTERGENIC | |
| SBS | Chr5 | 21535190 | G-T | HET | INTERGENIC | |
| SBS | Chr5 | 24451751 | G-C | HET | INTERGENIC | |
| SBS | Chr5 | 26406050 | G-A | HET | INTERGENIC | |
| SBS | Chr5 | 6038094 | C-A | HOMO | STOP_GAINED | LOC_Os05g10890 |
| SBS | Chr6 | 20009880 | C-T | HET | INTERGENIC | |
| SBS | Chr6 | 22125447 | A-G | HET | INTRON | |
| SBS | Chr6 | 30851725 | G-A | HET | INTERGENIC | |
| SBS | Chr6 | 5278420 | A-G | HOMO | INTRON | |
| SBS | Chr7 | 12351639 | T-A | HET | NON_SYNONYMOUS_CODING | LOC_Os07g22100 |
| SBS | Chr7 | 25487308 | T-A | HET | INTERGENIC | |
| SBS | Chr8 | 18529424 | G-A | HOMO | INTERGENIC | |
| SBS | Chr8 | 8146408 | A-G | HET | INTERGENIC | |
| SBS | Chr8 | 8324176 | T-C | HET | INTERGENIC | |
| SBS | Chr9 | 21823852 | A-G | HET | SYNONYMOUS_CODING | |
| SBS | Chr9 | 9469134 | G-A | HET | INTRON |
Deletions: 30
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr5 | 761146 | 761151 | 5 | LOC_Os05g02350 |
| Deletion | Chr5 | 2295098 | 2295100 | 2 | |
| Deletion | Chr2 | 5142113 | 5142127 | 14 | |
| Deletion | Chr1 | 5265401 | 5265412 | 11 | |
| Deletion | Chr3 | 5745217 | 5745220 | 3 | LOC_Os03g11140 |
| Deletion | Chr8 | 5753975 | 5753978 | 3 | |
| Deletion | Chr4 | 6678001 | 7183000 | 505000 | 77 |
| Deletion | Chr5 | 8517157 | 8517161 | 4 | LOC_Os05g15030 |
| Deletion | Chr1 | 8537901 | 8537902 | 1 | |
| Deletion | Chr11 | 9918086 | 9918091 | 5 | |
| Deletion | Chr11 | 11006491 | 11006493 | 2 | |
| Deletion | Chr3 | 12250949 | 12250957 | 8 | |
| Deletion | Chr6 | 13851109 | 13851119 | 10 | |
| Deletion | Chr12 | 15528279 | 15528698 | 419 | LOC_Os12g26560 |
| Deletion | Chr9 | 16660373 | 16660374 | 1 | |
| Deletion | Chr4 | 18189254 | 18189257 | 3 | |
| Deletion | Chr7 | 19011443 | 19011445 | 2 | |
| Deletion | Chr12 | 20728406 | 20728407 | 1 | |
| Deletion | Chr12 | 21248336 | 21248337 | 1 | LOC_Os12g34900 |
| Deletion | Chr2 | 23682021 | 23682022 | 1 | |
| Deletion | Chr1 | 24796682 | 24814493 | 17811 | 3 |
| Deletion | Chr5 | 25093144 | 25121429 | 28285 | LOC_Os05g43210 |
| Deletion | Chr8 | 26670360 | 26670361 | 1 | LOC_Os08g42230 |
| Deletion | Chr8 | 26689001 | 26708000 | 19000 | 5 |
| Deletion | Chr4 | 27274001 | 27274006 | 5 | |
| Deletion | Chr2 | 27658247 | 27658279 | 32 | |
| Deletion | Chr1 | 28084808 | 28084820 | 12 | |
| Deletion | Chr4 | 30735901 | 30735904 | 3 | |
| Deletion | Chr2 | 33251740 | 33251762 | 22 | |
| Deletion | Chr1 | 40107058 | 40107060 | 2 | LOC_Os01g69030 |
Insertions: 5
Inversions: 4
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr4 | 6678069 | 7292364 | LOC_Os04g12130 |
| Inversion | Chr4 | 7182634 | 7292363 | LOC_Os04g13020 |
| Inversion | Chr8 | 17685247 | 18482689 | 2 |
| Inversion | Chr8 | 17685276 | 18482707 | 2 |
Translocations: 4
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr2 | 7954255 | Chr1 | 15637525 | |
| Translocation | Chr8 | 20606601 | Chr1 | 27980090 | |
| Translocation | Chr11 | 23187882 | Chr4 | 16367928 | |
| Translocation | Chr4 | 30287048 | Chr3 | 744342 | LOC_Os03g02230 |
