Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN507-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN507-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 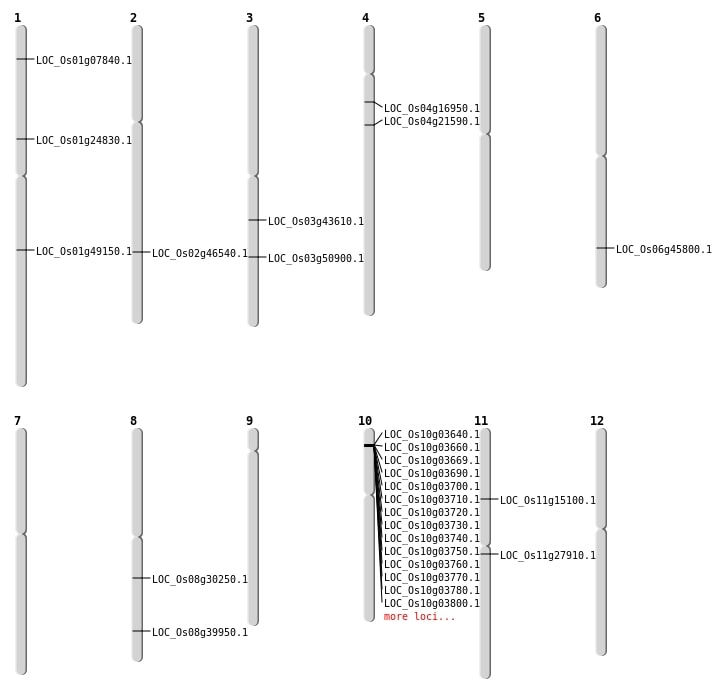
|
| Variant Information |
|---|
Single base substitutions: 35
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 13985196 | C-T | HOMO | NON_SYNONYMOUS_CODING | LOC_Os01g24830 |
| SBS | Chr1 | 3772026 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os01g07840 |
| SBS | Chr1 | 38478391 | A-T | HET | INTRON | |
| SBS | Chr1 | 4255063 | T-C | HOMO | SYNONYMOUS_CODING | |
| SBS | Chr1 | 5089683 | G-T | HOMO | INTERGENIC | |
| SBS | Chr10 | 15468601 | C-G | HOMO | INTERGENIC | |
| SBS | Chr10 | 1548295 | G-T | HOMO | INTERGENIC | |
| SBS | Chr10 | 1548297 | G-T | HOMO | INTERGENIC | |
| SBS | Chr11 | 22687044 | A-C | HET | INTERGENIC | |
| SBS | Chr11 | 8501259 | G-C | HET | NON_SYNONYMOUS_CODING | LOC_Os11g15100 |
| SBS | Chr11 | 8949681 | G-A | HOMO | SYNONYMOUS_CODING | |
| SBS | Chr12 | 4412438 | C-T | HET | INTERGENIC | |
| SBS | Chr2 | 12461393 | C-T | HET | INTERGENIC | |
| SBS | Chr2 | 12461394 | T-C | HET | INTERGENIC | |
| SBS | Chr2 | 28382762 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os02g46540 |
| SBS | Chr3 | 24374368 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os03g43610 |
| SBS | Chr3 | 32899611 | T-A | HET | INTERGENIC | |
| SBS | Chr4 | 12212612 | A-T | HET | NON_SYNONYMOUS_CODING | LOC_Os04g21590 |
| SBS | Chr4 | 14629365 | T-A | HOMO | INTERGENIC | |
| SBS | Chr4 | 15088155 | C-T | HET | INTRON | |
| SBS | Chr4 | 17468197 | C-T | HET | INTERGENIC | |
| SBS | Chr4 | 19204306 | T-C | HET | INTERGENIC | |
| SBS | Chr4 | 9284015 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os04g16950 |
| SBS | Chr5 | 12096744 | G-T | HET | INTRON | |
| SBS | Chr5 | 14171272 | G-A | HET | SYNONYMOUS_CODING | |
| SBS | Chr5 | 8539923 | C-A | HET | INTERGENIC | |
| SBS | Chr5 | 8664270 | C-T | HOMO | INTERGENIC | |
| SBS | Chr6 | 15158840 | C-A | HET | INTERGENIC | |
| SBS | Chr6 | 5482268 | G-A | HET | INTERGENIC | |
| SBS | Chr7 | 14001740 | A-G | HET | INTERGENIC | |
| SBS | Chr7 | 3245957 | G-T | HOMO | INTRON | |
| SBS | Chr8 | 11425651 | C-T | HET | INTERGENIC | |
| SBS | Chr8 | 18596181 | A-T | HET | NON_SYNONYMOUS_CODING | LOC_Os08g30250 |
| SBS | Chr8 | 18596182 | T-C | HET | SYNONYMOUS_CODING | |
| SBS | Chr9 | 15008630 | A-T | HET | INTERGENIC |
Deletions: 23
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr6 | 164311 | 164325 | 14 | |
| Deletion | Chr10 | 1626001 | 2746000 | 1120000 | 160 |
| Deletion | Chr8 | 2963971 | 2963974 | 3 | |
| Deletion | Chr9 | 4349164 | 4349165 | 1 | |
| Deletion | Chr3 | 6762215 | 6762217 | 2 | |
| Deletion | Chr11 | 7572396 | 7572402 | 6 | |
| Deletion | Chr11 | 7918873 | 7918888 | 15 | |
| Deletion | Chr9 | 8263827 | 8263829 | 2 | |
| Deletion | Chr9 | 8332705 | 8333009 | 304 | |
| Deletion | Chr3 | 9398895 | 9398902 | 7 | |
| Deletion | Chr10 | 9787506 | 9787516 | 10 | |
| Deletion | Chr2 | 12079171 | 12079172 | 1 | |
| Deletion | Chr9 | 15007250 | 15007262 | 12 | |
| Deletion | Chr4 | 15291163 | 15291164 | 1 | |
| Deletion | Chr11 | 16063217 | 16063223 | 6 | LOC_Os11g27910 |
| Deletion | Chr1 | 17243363 | 17243368 | 5 | |
| Deletion | Chr8 | 18896511 | 18896512 | 1 | |
| Deletion | Chr2 | 22523773 | 22523782 | 9 | |
| Deletion | Chr3 | 23716298 | 23716301 | 3 | |
| Deletion | Chr8 | 25313557 | 25313585 | 28 | LOC_Os08g39950 |
| Deletion | Chr6 | 27150999 | 27151003 | 4 | |
| Deletion | Chr6 | 27715316 | 27715331 | 15 | LOC_Os06g45800 |
| Deletion | Chr3 | 29079299 | 29079300 | 1 | LOC_Os03g50900 |
Insertions: 4
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr12 | 1472195 | 1472195 | 1 | |
| Insertion | Chr3 | 32775225 | 32775227 | 3 | |
| Insertion | Chr7 | 3753282 | 3753282 | 1 | |
| Insertion | Chr9 | 15024747 | 15024764 | 18 |
Inversions: 2
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr10 | 1589029 | 2746392 | 2 |
| Inversion | Chr11 | 20865402 | 28679726 |
Translocations: 3
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr10 | 2078914 | Chr7 | 29149114 | LOC_Os10g04400 |
| Translocation | Chr7 | 14001838 | Chr1 | 28244591 | LOC_Os01g49150 |
| Translocation | Chr9 | 20287708 | Chr6 | 22062273 |
