Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN512-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN512-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 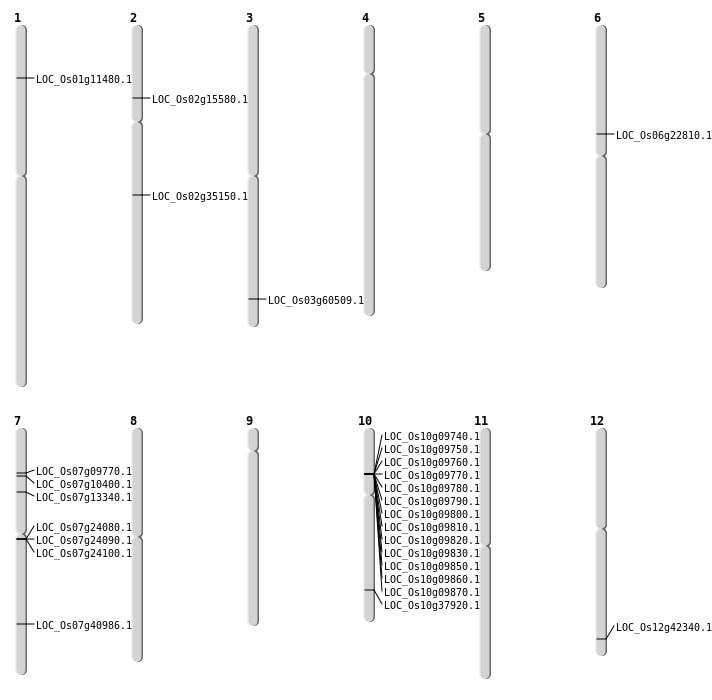
|
| Variant Information |
|---|
Single base substitutions: 44
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 13187750 | C-T | HET | INTERGENIC | |
| SBS | Chr1 | 29344350 | G-C | HOMO | INTERGENIC | |
| SBS | Chr1 | 3393632 | G-A | HOMO | INTERGENIC | |
| SBS | Chr10 | 13034035 | G-A | HET | INTERGENIC | |
| SBS | Chr10 | 14394005 | A-T | HET | INTERGENIC | |
| SBS | Chr10 | 20308300 | C-A | HET | SYNONYMOUS_CODING | |
| SBS | Chr10 | 20308301 | A-C | HET | NON_SYNONYMOUS_CODING | LOC_Os10g37920 |
| SBS | Chr10 | 3672465 | C-T | HET | INTERGENIC | |
| SBS | Chr11 | 18950872 | A-T | HET | INTERGENIC | |
| SBS | Chr11 | 18950873 | T-A | HET | INTERGENIC | |
| SBS | Chr11 | 21297944 | T-C | HET | SYNONYMOUS_CODING | |
| SBS | Chr11 | 3727284 | T-A | HET | INTERGENIC | |
| SBS | Chr11 | 9459651 | T-A | HET | INTERGENIC | |
| SBS | Chr2 | 18850283 | G-T | HOMO | INTRON | |
| SBS | Chr2 | 21112077 | T-A | HOMO | NON_SYNONYMOUS_CODING | LOC_Os02g35150 |
| SBS | Chr2 | 29691255 | A-G | HET | INTERGENIC | |
| SBS | Chr3 | 21554011 | C-T | HOMO | INTERGENIC | |
| SBS | Chr3 | 25275290 | C-T | HET | INTERGENIC | |
| SBS | Chr3 | 487107 | C-T | HOMO | INTERGENIC | |
| SBS | Chr3 | 532718 | A-T | HET | INTERGENIC | |
| SBS | Chr4 | 14859046 | C-T | HET | INTERGENIC | |
| SBS | Chr4 | 20788602 | A-G | HOMO | INTERGENIC | |
| SBS | Chr4 | 20788608 | T-G | HOMO | INTERGENIC | |
| SBS | Chr4 | 21840637 | A-C | HET | INTERGENIC | |
| SBS | Chr4 | 29802420 | T-A | HET | INTERGENIC | |
| SBS | Chr4 | 5155134 | A-C | HET | INTERGENIC | |
| SBS | Chr4 | 5997076 | T-A | HET | INTERGENIC | |
| SBS | Chr5 | 13629248 | A-C | HOMO | INTERGENIC | |
| SBS | Chr5 | 8823162 | G-A | HOMO | INTRON | |
| SBS | Chr6 | 13025327 | C-T | HOMO | INTERGENIC | |
| SBS | Chr6 | 25288518 | A-T | HET | INTERGENIC | |
| SBS | Chr6 | 30436203 | G-A | HOMO | INTRON | |
| SBS | Chr6 | 3801224 | T-C | HET | UTR_3_PRIME | |
| SBS | Chr7 | 10572017 | C-A | HET | INTRON | |
| SBS | Chr7 | 14637299 | G-T | HET | INTERGENIC | |
| SBS | Chr7 | 20783800 | G-A | HOMO | INTRON | |
| SBS | Chr7 | 4629321 | T-C | HET | INTRON | |
| SBS | Chr7 | 5160802 | G-C | HET | INTERGENIC | |
| SBS | Chr7 | 5206663 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os07g09770 |
| SBS | Chr7 | 8568985 | G-T | HET | INTERGENIC | |
| SBS | Chr8 | 12549247 | C-T | HET | INTERGENIC | |
| SBS | Chr8 | 13777220 | G-A | HOMO | SYNONYMOUS_CODING | |
| SBS | Chr9 | 16009483 | T-C | HET | INTERGENIC | |
| SBS | Chr9 | 20883495 | A-G | HET | INTERGENIC |
Deletions: 22
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr4 | 4940681 | 4940684 | 3 | |
| Deletion | Chr10 | 5267001 | 5341000 | 74000 | 13 |
| Deletion | Chr1 | 6182883 | 6185348 | 2465 | LOC_Os01g11480 |
| Deletion | Chr2 | 8754823 | 8754825 | 2 | LOC_Os02g15580 |
| Deletion | Chr8 | 11108537 | 11108538 | 1 | |
| Deletion | Chr12 | 13174230 | 13174233 | 3 | |
| Deletion | Chr6 | 13297173 | 13297178 | 5 | LOC_Os06g22810 |
| Deletion | Chr11 | 13631614 | 13631623 | 9 | |
| Deletion | Chr7 | 13651001 | 13662000 | 11000 | 3 |
| Deletion | Chr12 | 14638362 | 14638374 | 12 | |
| Deletion | Chr1 | 15642756 | 15642761 | 5 | |
| Deletion | Chr10 | 17933430 | 17933437 | 7 | |
| Deletion | Chr8 | 19238883 | 19238885 | 2 | |
| Deletion | Chr10 | 20894570 | 20894571 | 1 | |
| Deletion | Chr4 | 21736444 | 21736469 | 25 | |
| Deletion | Chr12 | 21916033 | 21916038 | 5 | |
| Deletion | Chr12 | 26326335 | 26326345 | 10 | LOC_Os12g42340 |
| Deletion | Chr7 | 26537476 | 26537477 | 1 | |
| Deletion | Chr6 | 28078797 | 28078798 | 1 | |
| Deletion | Chr4 | 30057564 | 30057588 | 24 | |
| Deletion | Chr4 | 33342906 | 33342907 | 1 | |
| Deletion | Chr1 | 38967436 | 38967437 | 1 |
Insertions: 3
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr1 | 11768578 | 11768579 | 2 | |
| Insertion | Chr3 | 6552857 | 6552857 | 1 | |
| Insertion | Chr6 | 8095788 | 8095788 | 1 |
Inversions: 3
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr7 | 5583804 | 7649894 | 2 |
| Inversion | Chr7 | 5627210 | 7649877 | LOC_Os07g13340 |
| Inversion | Chr6 | 22487425 | 22521001 |
Translocations: 5
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr10 | 5301381 | Chr6 | 14553623 | LOC_Os10g09800 |
| Translocation | Chr12 | 15184220 | Chr3 | 34394752 | LOC_Os03g60509 |
| Translocation | Chr9 | 21485600 | Chr7 | 24527375 | LOC_Os07g40986 |
| Translocation | Chr7 | 23795722 | Chr1 | 6185327 | |
| Translocation | Chr7 | 23795724 | Chr1 | 6182885 |
