Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN521-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN521-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 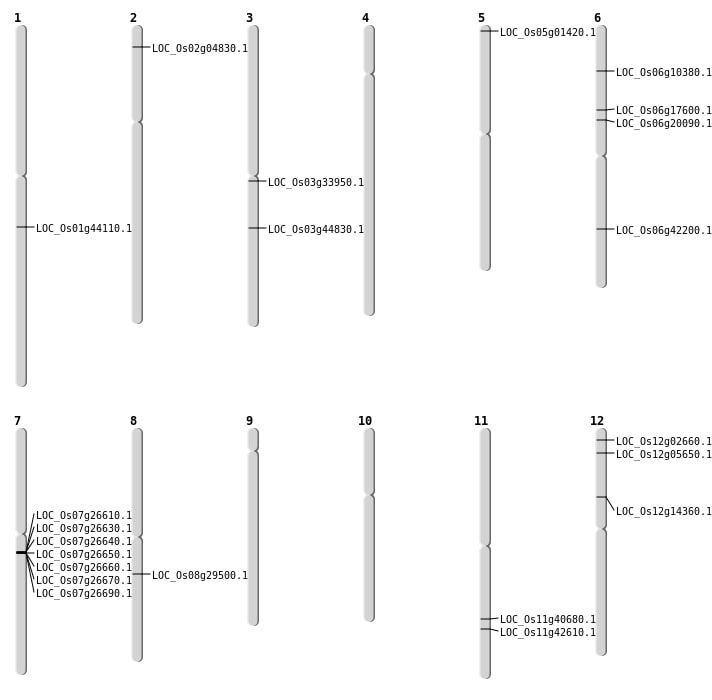
|
| Variant Information |
|---|
Single base substitutions: 41
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 15038411 | C-T | HET | INTERGENIC | |
| SBS | Chr1 | 20851564 | T-G | HET | INTRON | |
| SBS | Chr1 | 25098383 | C-T | HET | INTERGENIC | |
| SBS | Chr1 | 25098384 | A-T | HET | INTERGENIC | |
| SBS | Chr1 | 34539711 | T-C | HET | INTERGENIC | |
| SBS | Chr1 | 36381112 | T-C | HET | SYNONYMOUS_CODING | |
| SBS | Chr1 | 41402552 | T-C | HOMO | INTERGENIC | |
| SBS | Chr11 | 11616175 | C-T | HET | INTERGENIC | |
| SBS | Chr11 | 13579024 | G-A | HET | INTERGENIC | |
| SBS | Chr11 | 14800675 | G-A | HOMO | SYNONYMOUS_CODING | |
| SBS | Chr11 | 24279075 | A-G | HET | NON_SYNONYMOUS_CODING | LOC_Os11g40680 |
| SBS | Chr12 | 1323663 | G-A | HET | UTR_3_PRIME | |
| SBS | Chr12 | 19741952 | G-A | HET | INTRON | |
| SBS | Chr12 | 20409447 | G-A | HET | INTERGENIC | |
| SBS | Chr12 | 7720233 | G-A | HET | INTERGENIC | |
| SBS | Chr2 | 28690409 | A-T | HET | INTRON | |
| SBS | Chr3 | 15960352 | A-T | HET | INTERGENIC | |
| SBS | Chr3 | 32797005 | G-A | HET | INTERGENIC | |
| SBS | Chr4 | 10553685 | C-T | HOMO | INTRON | |
| SBS | Chr4 | 12807171 | C-A | HET | INTERGENIC | |
| SBS | Chr4 | 6871254 | G-A | HET | UTR_5_PRIME | |
| SBS | Chr5 | 15696575 | A-T | HET | INTERGENIC | |
| SBS | Chr5 | 15696576 | G-C | HET | INTERGENIC | |
| SBS | Chr5 | 1800351 | A-T | HET | INTERGENIC | |
| SBS | Chr5 | 20332219 | A-T | HET | INTERGENIC | |
| SBS | Chr5 | 5850668 | G-A | HET | INTERGENIC | |
| SBS | Chr5 | 6746149 | G-C | HET | INTERGENIC | |
| SBS | Chr6 | 25337731 | C-T | HOMO | NON_SYNONYMOUS_CODING | LOC_Os06g42200 |
| SBS | Chr6 | 375475 | A-G | HOMO | INTERGENIC | |
| SBS | Chr6 | 375480 | A-T | HOMO | INTERGENIC | |
| SBS | Chr6 | 5334656 | G-A | HOMO | SPLICE_SITE_DONOR | LOC_Os06g10380 |
| SBS | Chr6 | 5334657 | T-A | HOMO | SPLICE_SITE_DONOR | LOC_Os06g10380 |
| SBS | Chr7 | 12557332 | G-A | HOMO | INTERGENIC | |
| SBS | Chr7 | 16396100 | T-A | HOMO | INTERGENIC | |
| SBS | Chr7 | 6201360 | G-A | HET | SYNONYMOUS_CODING | |
| SBS | Chr7 | 6656326 | T-G | HET | INTERGENIC | |
| SBS | Chr7 | 718708 | A-T | HOMO | INTRON | |
| SBS | Chr8 | 18087953 | C-A | HET | NON_SYNONYMOUS_CODING | LOC_Os08g29500 |
| SBS | Chr8 | 18087954 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os08g29500 |
| SBS | Chr8 | 19543233 | G-A | HET | INTERGENIC | |
| SBS | Chr9 | 21496986 | C-T | HOMO | INTRON |
Deletions: 26
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr5 | 246629 | 246631 | 2 | LOC_Os05g01420 |
| Deletion | Chr11 | 3245519 | 3245520 | 1 | |
| Deletion | Chr9 | 4093645 | 4093654 | 9 | |
| Deletion | Chr8 | 4656179 | 4656184 | 5 | |
| Deletion | Chr9 | 5245417 | 5245418 | 1 | |
| Deletion | Chr6 | 5332880 | 5332885 | 5 | |
| Deletion | Chr12 | 8180665 | 8180674 | 9 | LOC_Os12g14360 |
| Deletion | Chr3 | 8975115 | 8975121 | 6 | |
| Deletion | Chr3 | 9379539 | 9379549 | 10 | |
| Deletion | Chr1 | 11116122 | 11116133 | 11 | |
| Deletion | Chr7 | 11336570 | 11336571 | 1 | |
| Deletion | Chr6 | 11515683 | 11515696 | 13 | LOC_Os06g20090 |
| Deletion | Chr3 | 13720592 | 13720595 | 3 | |
| Deletion | Chr2 | 14438182 | 14438185 | 3 | |
| Deletion | Chr7 | 15347584 | 15412589 | 65005 | 7 |
| Deletion | Chr7 | 16395912 | 16395914 | 2 | |
| Deletion | Chr6 | 18121443 | 18121444 | 1 | |
| Deletion | Chr1 | 19253859 | 19253860 | 1 | |
| Deletion | Chr11 | 22322410 | 22322415 | 5 | |
| Deletion | Chr1 | 25284937 | 25284949 | 12 | LOC_Os01g44110 |
| Deletion | Chr7 | 27184326 | 27184327 | 1 | |
| Deletion | Chr6 | 27710612 | 27710613 | 1 | |
| Deletion | Chr6 | 28336470 | 28336471 | 1 | |
| Deletion | Chr2 | 31914435 | 31914441 | 6 | |
| Deletion | Chr1 | 33155888 | 33155895 | 7 | |
| Deletion | Chr3 | 35886826 | 35886837 | 11 |
Insertions: 5
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr2 | 9518323 | 9518324 | 2 | |
| Insertion | Chr3 | 17060877 | 17060877 | 1 | |
| Insertion | Chr3 | 25279434 | 25279435 | 2 | LOC_Os03g44830 |
| Insertion | Chr5 | 1764989 | 1764991 | 3 | |
| Insertion | Chr8 | 27926856 | 27926860 | 5 |
Inversions: 1
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr2 | 473794 | 8683589 |
Translocations: 6
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr12 | 925307 | Chr11 | 9422842 | LOC_Os12g02660 |
| Translocation | Chr12 | 2597191 | Chr11 | 9422838 | LOC_Os12g05650 |
| Translocation | Chr9 | 11597066 | Chr6 | 10213552 | LOC_Os06g17600 |
| Translocation | Chr3 | 19375503 | Chr2 | 2226748 | 2 |
| Translocation | Chr4 | 24198576 | Chr1 | 2769632 | |
| Translocation | Chr11 | 25650376 | Chr7 | 9569351 | LOC_Os11g42610 |
