Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN522-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN522-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 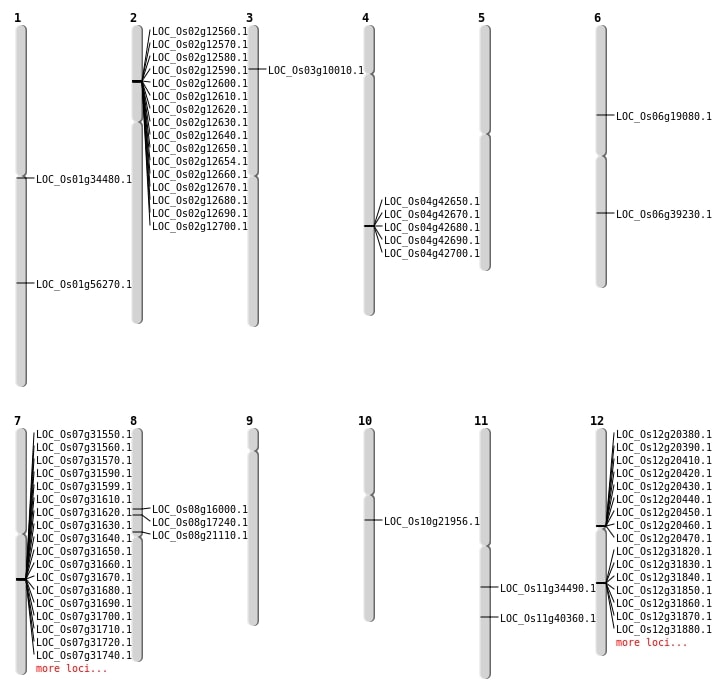
|
| Variant Information |
|---|
Single base substitutions: 33
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 12736041 | G-A | HOMO | INTRON | |
| SBS | Chr1 | 18139205 | C-T | HET | INTERGENIC | |
| SBS | Chr1 | 32411948 | A-G | HET | NON_SYNONYMOUS_CODING | LOC_Os01g56270 |
| SBS | Chr10 | 5530477 | C-T | HOMO | INTERGENIC | |
| SBS | Chr11 | 20206196 | A-C | HET | NON_SYNONYMOUS_CODING | LOC_Os11g34490 |
| SBS | Chr11 | 27309304 | G-T | HET | INTRON | |
| SBS | Chr12 | 11900711 | G-A | HET | INTERGENIC | |
| SBS | Chr12 | 16130069 | G-C | HET | INTRON | |
| SBS | Chr12 | 19145230 | T-A | HET | INTERGENIC | |
| SBS | Chr12 | 19145231 | T-A | HET | INTERGENIC | |
| SBS | Chr12 | 7780656 | C-T | HET | INTRON | |
| SBS | Chr2 | 18042347 | G-C | HOMO | INTRON | |
| SBS | Chr2 | 26026477 | A-C | HOMO | INTERGENIC | |
| SBS | Chr2 | 33032635 | G-T | HET | INTERGENIC | |
| SBS | Chr2 | 3756528 | C-G | HOMO | INTERGENIC | |
| SBS | Chr2 | 3756535 | A-T | HOMO | INTERGENIC | |
| SBS | Chr2 | 8375321 | G-T | HET | INTERGENIC | |
| SBS | Chr2 | 8375322 | A-T | HET | INTERGENIC | |
| SBS | Chr2 | 9446747 | T-A | HET | UTR_5_PRIME | |
| SBS | Chr3 | 13257973 | G-T | HET | INTERGENIC | |
| SBS | Chr3 | 2120939 | T-C | HET | INTRON | |
| SBS | Chr3 | 370488 | C-T | HET | SYNONYMOUS_CODING | |
| SBS | Chr3 | 5735209 | C-T | HET | INTRON | |
| SBS | Chr4 | 19456729 | C-T | HET | INTRON | |
| SBS | Chr5 | 19033293 | T-C | HET | INTERGENIC | |
| SBS | Chr6 | 17308617 | G-A | HET | INTERGENIC | |
| SBS | Chr6 | 8615563 | T-G | HET | INTERGENIC | |
| SBS | Chr8 | 10556310 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os08g17240 |
| SBS | Chr8 | 18971442 | C-T | HOMO | SYNONYMOUS_CODING | |
| SBS | Chr8 | 25197433 | G-A | HOMO | INTRON | |
| SBS | Chr8 | 25197434 | C-T | HOMO | INTRON | |
| SBS | Chr9 | 11733885 | C-T | HOMO | INTERGENIC | |
| SBS | Chr9 | 11733887 | G-T | HOMO | INTERGENIC |
Deletions: 23
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr8 | 2414889 | 2414896 | 7 | |
| Deletion | Chr12 | 4525404 | 4525411 | 7 | |
| Deletion | Chr3 | 5014647 | 5019248 | 4601 | LOC_Os03g10010 |
| Deletion | Chr5 | 6294019 | 6294028 | 9 | |
| Deletion | Chr8 | 6473467 | 6473486 | 19 | |
| Deletion | Chr2 | 6563001 | 6648000 | 85000 | 16 |
| Deletion | Chr10 | 7631084 | 7631085 | 1 | |
| Deletion | Chr3 | 9423463 | 9423464 | 1 | |
| Deletion | Chr8 | 9749953 | 9749957 | 4 | LOC_Os08g16000 |
| Deletion | Chr8 | 10821951 | 10821976 | 25 | |
| Deletion | Chr12 | 11901001 | 11957000 | 56000 | 8 |
| Deletion | Chr12 | 12269386 | 12269409 | 23 | |
| Deletion | Chr8 | 16522883 | 16522888 | 5 | |
| Deletion | Chr5 | 18436430 | 18436441 | 11 | |
| Deletion | Chr7 | 18731001 | 19031000 | 300000 | 38 |
| Deletion | Chr12 | 19149001 | 19301000 | 152000 | 18 |
| Deletion | Chr6 | 19578117 | 19578124 | 7 | |
| Deletion | Chr10 | 21574920 | 21574927 | 7 | |
| Deletion | Chr5 | 22669883 | 22669890 | 7 | |
| Deletion | Chr6 | 23291855 | 23291857 | 2 | LOC_Os06g39230 |
| Deletion | Chr11 | 24059964 | 24059980 | 16 | LOC_Os11g40360 |
| Deletion | Chr12 | 24291455 | 24292156 | 701 | 2 |
| Deletion | Chr4 | 25232001 | 25262000 | 30000 | 5 |
Insertions: 7
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr1 | 19009783 | 19009785 | 3 | LOC_Os01g34480 |
| Insertion | Chr11 | 13903938 | 13903940 | 3 | |
| Insertion | Chr3 | 29163199 | 29163199 | 1 | |
| Insertion | Chr4 | 21392479 | 21392480 | 2 | |
| Insertion | Chr6 | 10493040 | 10493041 | 2 | |
| Insertion | Chr9 | 4594876 | 4594877 | 2 | |
| Insertion | Chr9 | 5367792 | 5367792 | 1 |
Inversions: 2
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr8 | 10463201 | 12619219 | LOC_Os08g21110 |
| Inversion | Chr4 | 25187634 | 25286367 |
Translocations: 6
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr10 | 11338817 | Chr6 | 10850161 | 2 |
| Translocation | Chr12 | 11962593 | Chr9 | 11114431 | LOC_Os12g20470 |
| Translocation | Chr12 | 24128372 | Chr7 | 18730124 | LOC_Os07g31550 |
| Translocation | Chr12 | 24291455 | Chr3 | 5019297 | |
| Translocation | Chr12 | 24292152 | Chr4 | 25232112 | |
| Translocation | Chr4 | 25187713 | Chr2 | 10798269 |
