Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN524-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN524-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 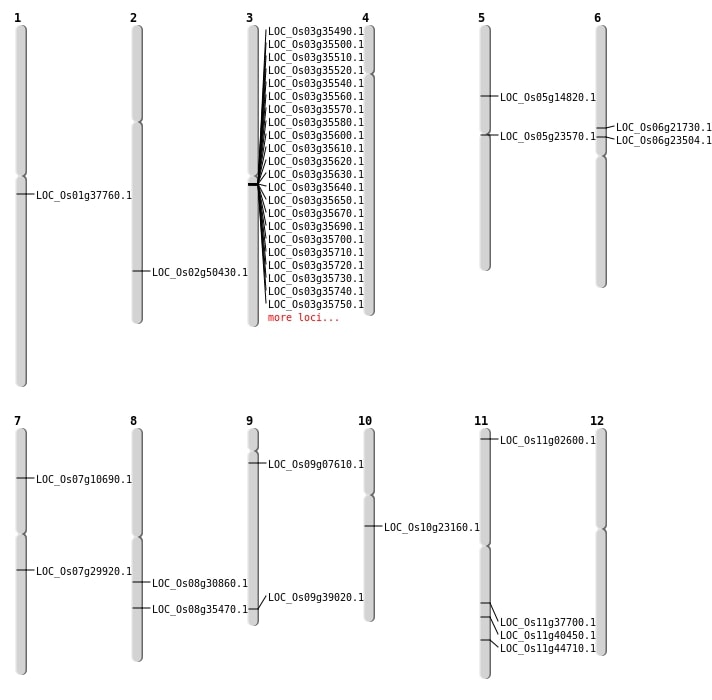
|
| Variant Information |
|---|
Single base substitutions: 34
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 14323390 | A-C | HOMO | INTERGENIC | |
| SBS | Chr1 | 4803652 | C-G | HET | INTERGENIC | |
| SBS | Chr1 | 8042262 | G-A | HET | INTERGENIC | |
| SBS | Chr10 | 14201591 | T-G | HET | INTERGENIC | |
| SBS | Chr10 | 16591985 | C-G | HET | INTERGENIC | |
| SBS | Chr10 | 18068753 | T-C | HET | INTERGENIC | |
| SBS | Chr10 | 4767354 | C-T | HET | SYNONYMOUS_CODING | |
| SBS | Chr10 | 8377240 | C-G | HOMO | INTRON | |
| SBS | Chr11 | 1067115 | G-A | HET | INTERGENIC | |
| SBS | Chr11 | 12019987 | C-T | HET | INTERGENIC | |
| SBS | Chr11 | 15962234 | G-A | HET | INTERGENIC | |
| SBS | Chr11 | 24125141 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os11g40450 |
| SBS | Chr11 | 27048225 | C-A | HET | NON_SYNONYMOUS_CODING | LOC_Os11g44710 |
| SBS | Chr11 | 7902282 | T-A | HET | INTERGENIC | |
| SBS | Chr11 | 8278287 | G-A | HET | INTERGENIC | |
| SBS | Chr12 | 7206074 | A-G | HET | INTERGENIC | |
| SBS | Chr12 | 960147 | C-T | HET | INTERGENIC | |
| SBS | Chr12 | 9640055 | G-T | HOMO | INTERGENIC | |
| SBS | Chr4 | 15242775 | G-A | HET | INTRON | |
| SBS | Chr4 | 18589590 | C-T | HET | INTERGENIC | |
| SBS | Chr5 | 6645786 | C-T | HET | INTERGENIC | |
| SBS | Chr6 | 13141259 | C-T | HET | INTERGENIC | |
| SBS | Chr6 | 15522066 | G-T | HET | INTERGENIC | |
| SBS | Chr6 | 2722477 | T-A | HOMO | INTERGENIC | |
| SBS | Chr6 | 7960298 | A-T | HET | INTERGENIC | |
| SBS | Chr6 | 8526061 | G-C | HET | INTERGENIC | |
| SBS | Chr7 | 10563308 | C-T | HOMO | INTERGENIC | |
| SBS | Chr7 | 10563309 | C-T | HOMO | INTERGENIC | |
| SBS | Chr7 | 25234811 | C-G | HET | INTERGENIC | |
| SBS | Chr7 | 555226 | T-C | HET | INTERGENIC | |
| SBS | Chr8 | 15826152 | C-G | HET | INTRON | |
| SBS | Chr8 | 19048966 | A-T | HET | NON_SYNONYMOUS_CODING | LOC_Os08g30860 |
| SBS | Chr9 | 16693233 | C-T | HET | INTERGENIC | |
| SBS | Chr9 | 2037727 | C-T | HET | INTERGENIC |
Deletions: 24
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr9 | 3823067 | 3823082 | 15 | LOC_Os09g07610 |
| Deletion | Chr5 | 5142488 | 5142496 | 8 | |
| Deletion | Chr7 | 5811492 | 5811545 | 53 | LOC_Os07g10690 |
| Deletion | Chr4 | 6365104 | 6365106 | 2 | |
| Deletion | Chr12 | 8388193 | 8388194 | 1 | |
| Deletion | Chr9 | 8873316 | 8873327 | 11 | |
| Deletion | Chr7 | 10563313 | 10563317 | 4 | |
| Deletion | Chr11 | 12679318 | 12679324 | 6 | |
| Deletion | Chr7 | 17608689 | 17621501 | 12812 | LOC_Os07g29920 |
| Deletion | Chr7 | 18996446 | 18996450 | 4 | |
| Deletion | Chr3 | 19673001 | 19882000 | 209000 | 27 |
| Deletion | Chr3 | 19884001 | 19976000 | 92000 | 15 |
| Deletion | Chr5 | 20772131 | 20772132 | 1 | |
| Deletion | Chr1 | 21056541 | 21056550 | 9 | |
| Deletion | Chr1 | 21123361 | 21123378 | 17 | LOC_Os01g37760 |
| Deletion | Chr11 | 22273337 | 22273338 | 1 | LOC_Os11g37700 |
| Deletion | Chr8 | 22355028 | 22355029 | 1 | LOC_Os08g35470 |
| Deletion | Chr9 | 22403749 | 22403774 | 25 | LOC_Os09g39020 |
| Deletion | Chr3 | 22408727 | 22408732 | 5 | |
| Deletion | Chr1 | 25122688 | 25122728 | 40 | |
| Deletion | Chr12 | 25255268 | 25255269 | 1 | |
| Deletion | Chr7 | 26816952 | 26816968 | 16 | LOC_Os07g44960 |
| Deletion | Chr2 | 30790997 | 30791006 | 9 | LOC_Os02g50430 |
| Deletion | Chr3 | 31285209 | 31285214 | 5 |
Insertions: 4
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr1 | 18826455 | 18826455 | 1 | |
| Insertion | Chr10 | 11958100 | 11958101 | 2 | |
| Insertion | Chr10 | 12086750 | 12086750 | 1 | LOC_Os10g23160 |
| Insertion | Chr8 | 24721344 | 24721345 | 2 |
Inversions: 3
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr4 | 4446145 | 16367899 | |
| Inversion | Chr6 | 12558377 | 13707621 | 2 |
| Inversion | Chr5 | 13517500 | 13701623 | LOC_Os05g23570 |
Translocations: 4
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr11 | 822540 | Chr4 | 16367929 | LOC_Os11g02600 |
| Translocation | Chr5 | 8428042 | Chr1 | 23528724 | LOC_Os05g14820 |
| Translocation | Chr11 | 25606195 | Chr7 | 26100972 | |
| Translocation | Chr11 | 25606198 | Chr7 | 26100846 |
