Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN525-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN525-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 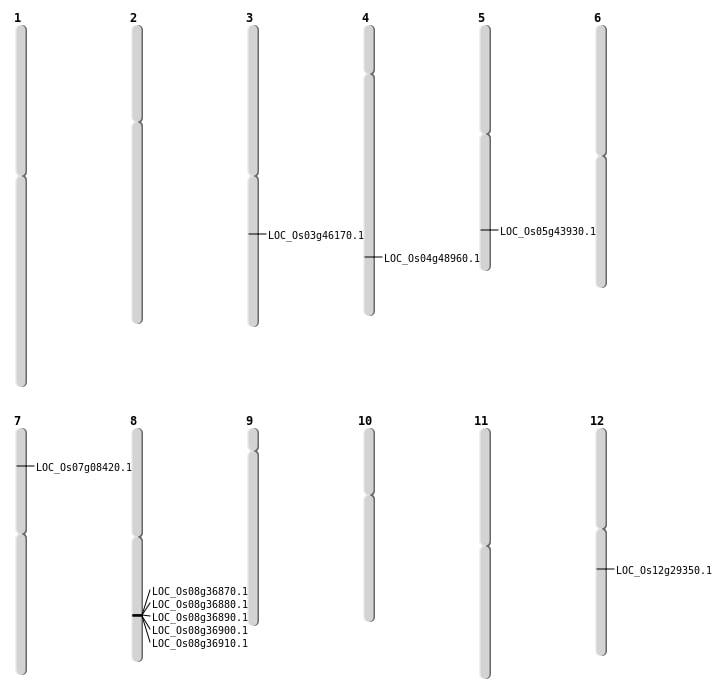
|
| Variant Information |
|---|
Single base substitutions: 21
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 12511647 | C-G | HOMO | INTRON | |
| SBS | Chr1 | 27435822 | T-C | HOMO | INTERGENIC | |
| SBS | Chr1 | 32885503 | G-A | HET | INTERGENIC | |
| SBS | Chr1 | 39765489 | C-T | HET | INTERGENIC | |
| SBS | Chr1 | 8914910 | C-T | HET | INTERGENIC | |
| SBS | Chr10 | 3889938 | C-T | HOMO | INTERGENIC | |
| SBS | Chr11 | 11566919 | C-A | HET | INTERGENIC | |
| SBS | Chr11 | 13788215 | T-A | HET | INTERGENIC | |
| SBS | Chr12 | 22723359 | A-G | HET | UTR_3_PRIME | |
| SBS | Chr12 | 23814415 | T-A | HET | INTERGENIC | |
| SBS | Chr2 | 17679326 | A-C | HOMO | INTERGENIC | |
| SBS | Chr2 | 33452980 | G-T | HET | INTERGENIC | |
| SBS | Chr2 | 33452981 | G-T | HET | INTERGENIC | |
| SBS | Chr3 | 26103460 | T-G | HET | NON_SYNONYMOUS_CODING | LOC_Os03g46170 |
| SBS | Chr3 | 329969 | T-C | HET | INTERGENIC | |
| SBS | Chr4 | 25264511 | C-T | HET | SYNONYMOUS_CODING | |
| SBS | Chr7 | 15987347 | G-A | HET | INTERGENIC | |
| SBS | Chr7 | 20650492 | C-A | HOMO | INTRON | |
| SBS | Chr8 | 23704206 | T-A | HOMO | INTERGENIC | |
| SBS | Chr8 | 27000297 | A-T | HET | INTERGENIC | |
| SBS | Chr9 | 2416264 | A-G | HOMO | INTRON |
Deletions: 21
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr5 | 631033 | 631044 | 11 | |
| Deletion | Chr9 | 4206960 | 4206961 | 1 | |
| Deletion | Chr7 | 4326357 | 4326358 | 1 | LOC_Os07g08420 |
| Deletion | Chr11 | 4410388 | 4410390 | 2 | |
| Deletion | Chr10 | 5563003 | 5563008 | 5 | |
| Deletion | Chr1 | 5730976 | 5731107 | 131 | |
| Deletion | Chr2 | 8441428 | 8441437 | 9 | |
| Deletion | Chr9 | 8476625 | 8476629 | 4 | |
| Deletion | Chr10 | 8914072 | 8914079 | 7 | |
| Deletion | Chr8 | 9338440 | 9338441 | 1 | |
| Deletion | Chr8 | 11067511 | 11067518 | 7 | |
| Deletion | Chr6 | 15148899 | 15148908 | 9 | |
| Deletion | Chr7 | 16844771 | 16844778 | 7 | |
| Deletion | Chr12 | 17420445 | 17420454 | 9 | LOC_Os12g29350 |
| Deletion | Chr6 | 17763099 | 17763100 | 1 | |
| Deletion | Chr5 | 21363945 | 21363948 | 3 | |
| Deletion | Chr1 | 21914645 | 21914646 | 1 | |
| Deletion | Chr8 | 23296001 | 23344000 | 48000 | 5 |
| Deletion | Chr4 | 23572747 | 23572758 | 11 | |
| Deletion | Chr5 | 25564206 | 25569080 | 4874 | LOC_Os05g43930 |
| Deletion | Chr1 | 29683880 | 29683881 | 1 |
Insertions: 4
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr1 | 8930793 | 8930795 | 3 | |
| Insertion | Chr10 | 11739229 | 11739230 | 2 | |
| Insertion | Chr7 | 20126629 | 20126630 | 2 | |
| Insertion | Chr7 | 2174770 | 2174770 | 1 |
No Inversion
Translocations: 6
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr12 | 6993247 | Chr3 | 22046065 | |
| Translocation | Chr8 | 14785729 | Chr1 | 20577303 | |
| Translocation | Chr6 | 22819227 | Chr4 | 29199076 | LOC_Os04g48960 |
| Translocation | Chr6 | 22949840 | Chr4 | 29199069 | LOC_Os04g48960 |
| Translocation | Chr5 | 25564240 | Chr2 | 29257101 | |
| Translocation | Chr5 | 25569071 | Chr2 | 29257060 |
