Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN530-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN530-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 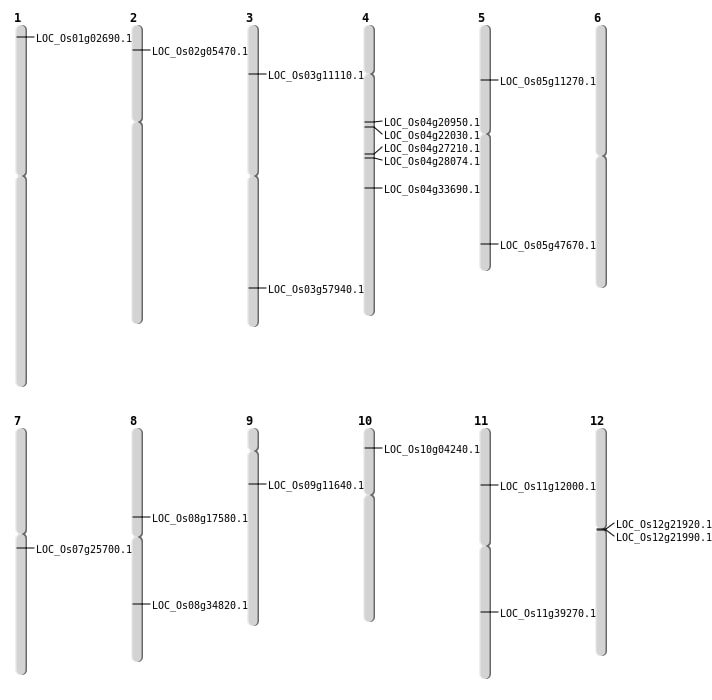
|
| Variant Information |
|---|
Single base substitutions: 31
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 43140227 | A-G | HET | INTERGENIC | |
| SBS | Chr10 | 1974669 | C-T | HOMO | NON_SYNONYMOUS_CODING | LOC_Os10g04240 |
| SBS | Chr11 | 1062294 | A-C | HET | INTERGENIC | |
| SBS | Chr12 | 12371239 | A-G | HOMO | NON_SYNONYMOUS_CODING | LOC_Os12g21990 |
| SBS | Chr12 | 15920654 | T-G | HET | INTERGENIC | |
| SBS | Chr2 | 18702235 | G-A | HET | INTERGENIC | |
| SBS | Chr2 | 27050609 | T-A | HET | INTRON | |
| SBS | Chr2 | 35413813 | C-A | HOMO | INTERGENIC | |
| SBS | Chr3 | 11245927 | T-C | HET | INTRON | |
| SBS | Chr3 | 22038798 | G-T | HET | INTERGENIC | |
| SBS | Chr4 | 11142254 | G-T | HOMO | INTRON | |
| SBS | Chr4 | 11759723 | C-T | HOMO | NON_SYNONYMOUS_CODING | LOC_Os04g20950 |
| SBS | Chr4 | 12478871 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os04g22030 |
| SBS | Chr4 | 22533771 | G-A | HOMO | INTERGENIC | |
| SBS | Chr4 | 2361885 | C-A | HET | SYNONYMOUS_CODING | |
| SBS | Chr4 | 25430733 | A-T | HET | INTERGENIC | |
| SBS | Chr4 | 25668861 | G-T | HET | INTERGENIC | |
| SBS | Chr4 | 7279312 | G-A | HOMO | INTRON | |
| SBS | Chr5 | 27317756 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os05g47670 |
| SBS | Chr5 | 27317757 | G-A | HET | SYNONYMOUS_CODING | |
| SBS | Chr5 | 29484051 | T-C | HET | INTRON | |
| SBS | Chr5 | 4882662 | T-A | HET | INTRON | |
| SBS | Chr6 | 11569199 | G-A | HOMO | SYNONYMOUS_CODING | |
| SBS | Chr6 | 23187259 | C-A | HOMO | UTR_3_PRIME | |
| SBS | Chr6 | 23187260 | G-A | HOMO | UTR_3_PRIME | |
| SBS | Chr6 | 29862591 | C-T | HET | INTERGENIC | |
| SBS | Chr7 | 14742841 | A-G | HOMO | NON_SYNONYMOUS_CODING | LOC_Os07g25700 |
| SBS | Chr7 | 15592494 | G-A | HOMO | INTERGENIC | |
| SBS | Chr8 | 21906220 | C-T | HOMO | NON_SYNONYMOUS_CODING | LOC_Os08g34820 |
| SBS | Chr8 | 26738259 | A-C | HET | INTERGENIC | |
| SBS | Chr9 | 6092927 | A-G | HET | INTERGENIC |
Deletions: 25
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr2 | 2641539 | 2641589 | 50 | LOC_Os02g05470 |
| Deletion | Chr8 | 4249145 | 4249147 | 2 | |
| Deletion | Chr10 | 4488955 | 4488965 | 10 | |
| Deletion | Chr6 | 4645457 | 4645463 | 6 | |
| Deletion | Chr1 | 4657342 | 4657344 | 2 | |
| Deletion | Chr1 | 4878534 | 4878539 | 5 | |
| Deletion | Chr3 | 5714918 | 5714919 | 1 | LOC_Os03g11110 |
| Deletion | Chr9 | 6498562 | 6498564 | 2 | LOC_Os09g11640 |
| Deletion | Chr10 | 8821758 | 8821767 | 9 | |
| Deletion | Chr4 | 8917566 | 8917571 | 5 | |
| Deletion | Chr12 | 12345386 | 12345400 | 14 | LOC_Os12g21920 |
| Deletion | Chr3 | 16568831 | 16568835 | 4 | |
| Deletion | Chr3 | 16710204 | 16710211 | 7 | |
| Deletion | Chr12 | 17634695 | 17634697 | 2 | |
| Deletion | Chr8 | 17986504 | 17986522 | 18 | |
| Deletion | Chr3 | 18065264 | 18065275 | 11 | |
| Deletion | Chr9 | 19464858 | 19464872 | 14 | |
| Deletion | Chr8 | 19944937 | 19944942 | 5 | |
| Deletion | Chr4 | 20398277 | 20398278 | 1 | LOC_Os04g33690 |
| Deletion | Chr7 | 20810998 | 20810999 | 1 | |
| Deletion | Chr6 | 23179879 | 23179890 | 11 | |
| Deletion | Chr5 | 25154711 | 25154725 | 14 | |
| Deletion | Chr5 | 26693218 | 26693220 | 2 | |
| Deletion | Chr3 | 33002893 | 33002904 | 11 | LOC_Os03g57940 |
| Deletion | Chr3 | 36070555 | 36070556 | 1 |
Insertions: 7
Inversions: 2
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr1 | 918002 | 2717377 | LOC_Os01g02690 |
| Inversion | Chr1 | 918014 | 2717385 | LOC_Os01g02690 |
Translocations: 6
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr5 | 6366367 | Chr4 | 16083641 | 2 |
| Translocation | Chr11 | 6675672 | Chr5 | 19243485 | LOC_Os11g12000 |
| Translocation | Chr8 | 10754437 | Chr6 | 19260574 | LOC_Os08g17580 |
| Translocation | Chr4 | 16578611 | Chr1 | 36180932 | LOC_Os04g28074 |
| Translocation | Chr11 | 23382761 | Chr5 | 16792647 | LOC_Os11g39270 |
| Translocation | Chr11 | 23382764 | Chr5 | 16789341 | LOC_Os11g39270 |
