Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN548-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN548-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 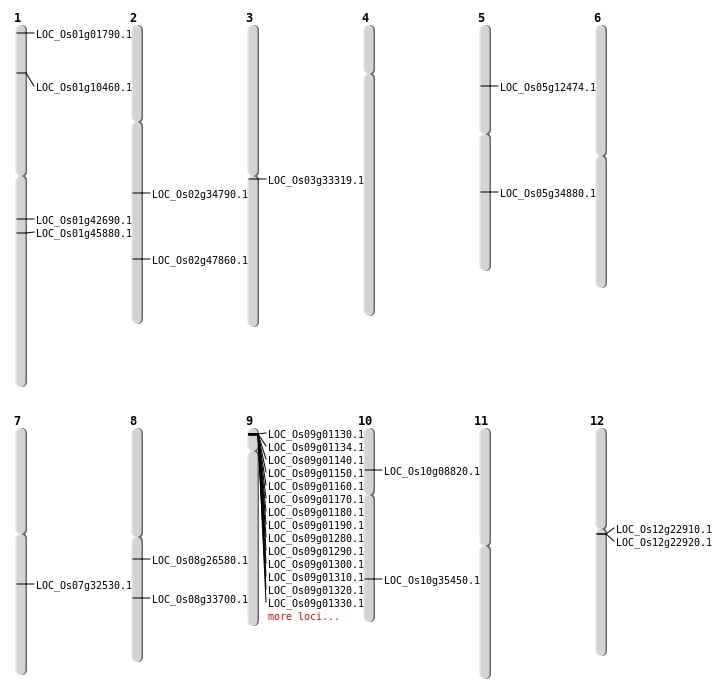
|
| Variant Information |
|---|
Single base substitutions: 33
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 16112739 | A-G | HOMO | INTERGENIC | |
| SBS | Chr1 | 1792318 | A-T | HET | INTERGENIC | |
| SBS | Chr1 | 31390654 | T-A | HET | INTERGENIC | |
| SBS | Chr1 | 37282772 | A-G | HET | INTRON | |
| SBS | Chr1 | 42324700 | G-A | HET | INTERGENIC | |
| SBS | Chr1 | 42324701 | T-A | HET | INTERGENIC | |
| SBS | Chr1 | 9527394 | A-T | HET | INTERGENIC | |
| SBS | Chr10 | 18973769 | C-T | HOMO | NON_SYNONYMOUS_CODING | LOC_Os10g35450 |
| SBS | Chr11 | 13367542 | C-T | HOMO | INTERGENIC | |
| SBS | Chr11 | 16971935 | G-A | HOMO | INTRON | |
| SBS | Chr12 | 20802048 | T-A | HOMO | SPLICE_SITE_REGION | |
| SBS | Chr12 | 26825818 | A-T | HET | INTRON | |
| SBS | Chr12 | 4610690 | T-A | HET | INTERGENIC | |
| SBS | Chr2 | 23662535 | G-A | HET | INTERGENIC | |
| SBS | Chr2 | 27673156 | T-A | HET | INTERGENIC | |
| SBS | Chr2 | 27673157 | C-A | HET | INTERGENIC | |
| SBS | Chr3 | 25219144 | T-C | HOMO | INTERGENIC | |
| SBS | Chr3 | 30413631 | C-T | HET | INTERGENIC | |
| SBS | Chr3 | 31658116 | C-T | HET | UTR_3_PRIME | |
| SBS | Chr4 | 23338071 | G-T | HOMO | INTERGENIC | |
| SBS | Chr4 | 9756003 | G-A | HOMO | INTERGENIC | |
| SBS | Chr5 | 19521221 | G-C | HET | INTERGENIC | |
| SBS | Chr6 | 13571314 | T-C | HET | INTERGENIC | |
| SBS | Chr6 | 13571317 | A-T | HET | INTERGENIC | |
| SBS | Chr6 | 22417130 | G-A | HET | SYNONYMOUS_CODING | |
| SBS | Chr6 | 2581039 | A-T | HOMO | INTRON | |
| SBS | Chr7 | 6509764 | C-A | HOMO | INTERGENIC | |
| SBS | Chr8 | 11275142 | C-G | HET | UTR_3_PRIME | |
| SBS | Chr8 | 2596687 | C-T | HET | INTERGENIC | |
| SBS | Chr8 | 3082543 | C-A | HET | INTRON | |
| SBS | Chr9 | 12775471 | C-A | HOMO | INTERGENIC | |
| SBS | Chr9 | 4288697 | C-T | HET | INTERGENIC | |
| SBS | Chr9 | 5709940 | A-G | HET | SYNONYMOUS_CODING |
Deletions: 28
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr9 | 134001 | 178000 | 44000 | 8 |
| Deletion | Chr9 | 240001 | 304000 | 64000 | 9 |
| Deletion | Chr5 | 2176412 | 2176413 | 1 | |
| Deletion | Chr4 | 2723994 | 2723995 | 1 | |
| Deletion | Chr10 | 4772011 | 4772012 | 1 | LOC_Os10g08820 |
| Deletion | Chr3 | 4928481 | 4928482 | 1 | |
| Deletion | Chr7 | 6515583 | 6515609 | 26 | |
| Deletion | Chr5 | 7168146 | 7168156 | 10 | LOC_Os05g12474 |
| Deletion | Chr6 | 7592393 | 7592404 | 11 | |
| Deletion | Chr4 | 9245596 | 9245603 | 7 | |
| Deletion | Chr6 | 12260373 | 12260374 | 1 | |
| Deletion | Chr5 | 12427154 | 12427177 | 23 | |
| Deletion | Chr3 | 12899712 | 12899713 | 1 | |
| Deletion | Chr12 | 12935001 | 12952000 | 17000 | 2 |
| Deletion | Chr8 | 13118120 | 13118121 | 1 | |
| Deletion | Chr3 | 19057186 | 19057199 | 13 | LOC_Os03g33319 |
| Deletion | Chr7 | 19385936 | 19385944 | 8 | LOC_Os07g32530 |
| Deletion | Chr1 | 20080667 | 20080671 | 4 | |
| Deletion | Chr2 | 20858611 | 20858617 | 6 | LOC_Os02g34790 |
| Deletion | Chr8 | 21045493 | 21045505 | 12 | LOC_Os08g33700 |
| Deletion | Chr3 | 22840332 | 22840334 | 2 | |
| Deletion | Chr1 | 24013203 | 24013205 | 2 | |
| Deletion | Chr1 | 24279828 | 24279897 | 69 | LOC_Os01g42690 |
| Deletion | Chr2 | 25212323 | 25212327 | 4 | |
| Deletion | Chr2 | 26238371 | 26238374 | 3 | |
| Deletion | Chr7 | 28552348 | 28552349 | 1 | |
| Deletion | Chr1 | 28942355 | 28942365 | 10 | |
| Deletion | Chr2 | 29281482 | 29281492 | 10 | LOC_Os02g47860 |
Insertions: 5
Inversions: 4
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr1 | 420047 | 617582 | LOC_Os01g01790 |
| Inversion | Chr9 | 436035 | 3487949 | LOC_Os09g07140 |
| Inversion | Chr9 | 436209 | 3487946 | LOC_Os09g07140 |
| Inversion | Chr1 | 1681132 | 5533479 | LOC_Os01g10460 |
Translocations: 5
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr9 | 1863445 | Chr1 | 26061941 | 2 |
| Translocation | Chr3 | 11169112 | Chr2 | 34742296 | |
| Translocation | Chr10 | 15993807 | Chr8 | 16179600 | LOC_Os08g26580 |
| Translocation | Chr5 | 20725553 | Chr2 | 5558257 | LOC_Os05g34880 |
| Translocation | Chr7 | 24230191 | Chr4 | 18481655 |
