Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN576-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN576-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 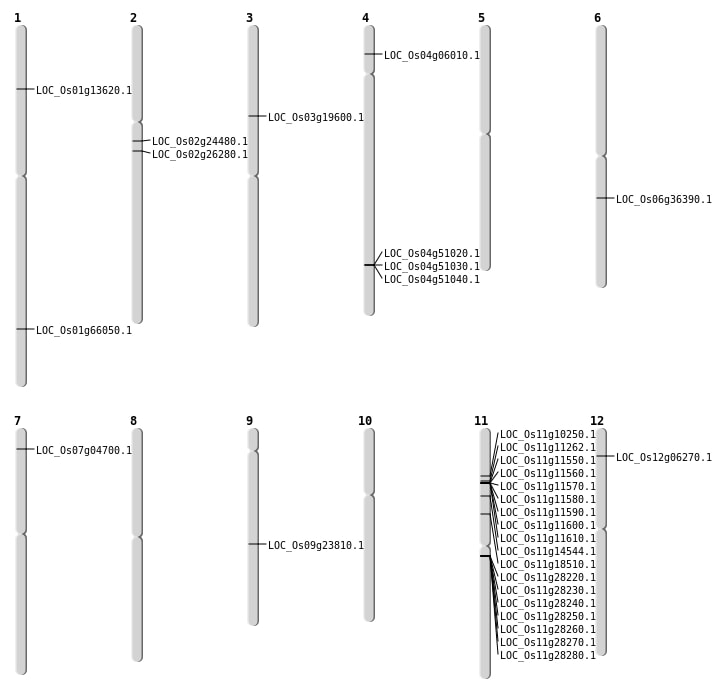
|
| Variant Information |
|---|
Single base substitutions: 40
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 5920304 | A-G | HET | INTERGENIC | |
| SBS | Chr1 | 7008403 | G-T | HOMO | INTERGENIC | |
| SBS | Chr1 | 7640625 | C-G | HET | NON_SYNONYMOUS_CODING | LOC_Os01g13620 |
| SBS | Chr1 | 7852020 | A-T | HET | INTERGENIC | |
| SBS | Chr10 | 12654956 | A-G | HOMO | INTERGENIC | |
| SBS | Chr10 | 586034 | C-A | HOMO | INTERGENIC | |
| SBS | Chr10 | 7525217 | T-C | HOMO | INTRON | |
| SBS | Chr11 | 10435995 | T-C | HET | NON_SYNONYMOUS_CODING | LOC_Os11g18510 |
| SBS | Chr11 | 17206488 | G-A | HET | INTERGENIC | |
| SBS | Chr11 | 20984773 | C-G | HET | INTERGENIC | |
| SBS | Chr12 | 11468824 | T-G | HOMO | UTR_3_PRIME | |
| SBS | Chr12 | 19977282 | A-G | HET | INTRON | |
| SBS | Chr12 | 2745916 | G-A | HOMO | INTERGENIC | |
| SBS | Chr2 | 14180966 | C-G | HET | NON_SYNONYMOUS_CODING | LOC_Os02g24480 |
| SBS | Chr2 | 15430986 | G-T | HOMO | NON_SYNONYMOUS_CODING | LOC_Os02g26280 |
| SBS | Chr2 | 15553281 | C-T | HOMO | INTRON | |
| SBS | Chr2 | 15553290 | A-G | HOMO | INTRON | |
| SBS | Chr3 | 11032294 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os03g19600 |
| SBS | Chr3 | 1843288 | G-T | HET | INTERGENIC | |
| SBS | Chr3 | 1843289 | A-T | HET | INTERGENIC | |
| SBS | Chr3 | 22939602 | C-T | HOMO | INTERGENIC | |
| SBS | Chr3 | 31783985 | A-T | HOMO | INTERGENIC | |
| SBS | Chr4 | 16315089 | G-A | HOMO | INTERGENIC | |
| SBS | Chr4 | 18382055 | C-T | HET | INTERGENIC | |
| SBS | Chr4 | 26353955 | T-C | HET | INTERGENIC | |
| SBS | Chr4 | 3116779 | C-T | HOMO | STOP_GAINED | LOC_Os04g06010 |
| SBS | Chr4 | 5042545 | A-G | HET | SPLICE_SITE_REGION | |
| SBS | Chr5 | 14832644 | C-T | HOMO | INTERGENIC | |
| SBS | Chr5 | 19309717 | G-A | HOMO | INTERGENIC | |
| SBS | Chr5 | 3519185 | G-T | HOMO | INTERGENIC | |
| SBS | Chr6 | 20104567 | C-T | HET | INTERGENIC | |
| SBS | Chr6 | 669955 | G-A | HET | INTERGENIC | |
| SBS | Chr7 | 5539266 | A-C | HET | INTERGENIC | |
| SBS | Chr7 | 7126831 | G-A | HET | INTERGENIC | |
| SBS | Chr7 | 7665599 | T-C | HET | INTERGENIC | |
| SBS | Chr8 | 14275996 | C-T | HOMO | INTERGENIC | |
| SBS | Chr8 | 7752170 | T-C | HET | INTERGENIC | |
| SBS | Chr9 | 14188553 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os09g23810 |
| SBS | Chr9 | 20654288 | G-A | HET | INTERGENIC | |
| SBS | Chr9 | 7082778 | T-G | HET | INTERGENIC |
Deletions: 19
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr11 | 768064 | 768069 | 5 | |
| Deletion | Chr10 | 1508141 | 1508143 | 2 | |
| Deletion | Chr7 | 2084356 | 2084362 | 6 | LOC_Os07g04700 |
| Deletion | Chr3 | 2522886 | 2522896 | 10 | |
| Deletion | Chr7 | 3283309 | 3283314 | 5 | |
| Deletion | Chr11 | 6424001 | 6458000 | 34000 | 7 |
| Deletion | Chr11 | 8162152 | 8163431 | 1279 | LOC_Os11g14544 |
| Deletion | Chr1 | 8218110 | 8218410 | 300 | |
| Deletion | Chr7 | 9012422 | 9012430 | 8 | |
| Deletion | Chr6 | 12289286 | 12289288 | 2 | |
| Deletion | Chr1 | 12431652 | 12431653 | 1 | |
| Deletion | Chr11 | 16221001 | 16273000 | 52000 | 7 |
| Deletion | Chr4 | 16294617 | 16294619 | 2 | |
| Deletion | Chr5 | 24347445 | 24347446 | 1 | |
| Deletion | Chr8 | 27883292 | 27883328 | 36 | |
| Deletion | Chr4 | 28953259 | 28953299 | 40 | |
| Deletion | Chr3 | 29097674 | 29097679 | 5 | |
| Deletion | Chr4 | 30201756 | 30219650 | 17894 | 3 |
| Deletion | Chr1 | 38330224 | 38330234 | 10 | LOC_Os01g66050 |
Insertions: 4
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr4 | 28175908 | 28175911 | 4 | |
| Insertion | Chr5 | 18183649 | 18183649 | 1 | |
| Insertion | Chr5 | 4779596 | 4779597 | 2 | |
| Insertion | Chr6 | 21372446 | 21372451 | 6 | LOC_Os06g36390 |
Inversions: 5
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr12 | 1772343 | 2990107 | LOC_Os12g06270 |
| Inversion | Chr11 | 5539100 | 6233776 | LOC_Os11g11262 |
| Inversion | Chr11 | 5565150 | 6470917 | LOC_Os11g10250 |
| Inversion | Chr11 | 5566656 | 6232581 | LOC_Os11g10250 |
| Inversion | Chr11 | 6233761 | 6423248 | 2 |
Translocations: 2
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr12 | 1772394 | Chr1 | 8218444 | |
| Translocation | Chr12 | 2992491 | Chr1 | 8218131 | LOC_Os12g06270 |
