Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN579-S [Download] |
| Generation | M2-SOS |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN579-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 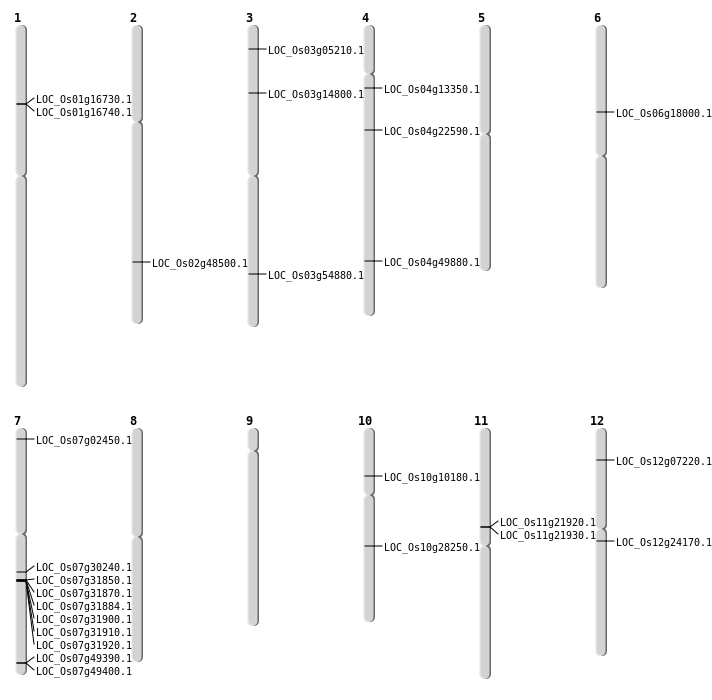
|
| Variant Information |
|---|
Single base substitutions: 38
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 34284916 | C-T | HET | INTERGENIC | |
| SBS | Chr1 | 40737317 | G-T | HOMO | INTERGENIC | |
| SBS | Chr1 | 5906184 | G-A | HET | UTR_5_PRIME | |
| SBS | Chr10 | 12243697 | C-T | HOMO | INTERGENIC | |
| SBS | Chr11 | 14666749 | C-T | HOMO | INTRON | |
| SBS | Chr11 | 25202320 | C-T | HOMO | INTRON | |
| SBS | Chr12 | 10119252 | G-C | HET | INTERGENIC | |
| SBS | Chr12 | 10998032 | C-T | HOMO | INTERGENIC | |
| SBS | Chr12 | 12255365 | G-C | HOMO | INTRON | |
| SBS | Chr12 | 23544543 | C-A | HOMO | INTERGENIC | |
| SBS | Chr12 | 25328360 | T-C | HOMO | INTRON | |
| SBS | Chr12 | 3550366 | T-G | HET | NON_SYNONYMOUS_CODING | LOC_Os12g07220 |
| SBS | Chr2 | 11144247 | G-A | HOMO | INTRON | |
| SBS | Chr2 | 30858489 | C-T | HET | INTERGENIC | |
| SBS | Chr3 | 19677465 | C-A | HET | INTERGENIC | |
| SBS | Chr3 | 2530312 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os03g05210 |
| SBS | Chr3 | 26301883 | G-T | HOMO | INTERGENIC | |
| SBS | Chr3 | 26301884 | C-T | HOMO | INTERGENIC | |
| SBS | Chr3 | 2737460 | C-A | HET | INTERGENIC | |
| SBS | Chr3 | 31198028 | A-T | HET | START_LOST | LOC_Os03g54880 |
| SBS | Chr4 | 12791442 | C-T | HOMO | NON_SYNONYMOUS_CODING | LOC_Os04g22590 |
| SBS | Chr4 | 29740116 | C-T | HOMO | NON_SYNONYMOUS_CODING | LOC_Os04g49880 |
| SBS | Chr5 | 15008004 | C-T | HET | INTERGENIC | |
| SBS | Chr5 | 17022420 | T-A | HET | INTERGENIC | |
| SBS | Chr5 | 17350140 | G-A | HET | INTERGENIC | |
| SBS | Chr5 | 22217573 | T-C | HOMO | INTRON | |
| SBS | Chr6 | 17464669 | G-A | HET | INTERGENIC | |
| SBS | Chr6 | 5178896 | G-C | HET | INTERGENIC | |
| SBS | Chr6 | 5459841 | C-G | HET | INTERGENIC | |
| SBS | Chr6 | 9904842 | G-A | HET | INTRON | |
| SBS | Chr7 | 18367356 | G-A | HOMO | INTERGENIC | |
| SBS | Chr7 | 843760 | C-G | HOMO | NON_SYNONYMOUS_CODING | LOC_Os07g02450 |
| SBS | Chr8 | 10692829 | G-A | HOMO | INTRON | |
| SBS | Chr8 | 14215141 | G-A | HET | INTRON | |
| SBS | Chr8 | 15708588 | G-A | HOMO | INTERGENIC | |
| SBS | Chr8 | 7267425 | C-T | HOMO | INTERGENIC | |
| SBS | Chr9 | 461048 | G-A | HOMO | INTERGENIC | |
| SBS | Chr9 | 721378 | G-A | HET | INTERGENIC |
Deletions: 24
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr10 | 4669914 | 4669919 | 5 | |
| Deletion | Chr5 | 5207922 | 5207924 | 2 | |
| Deletion | Chr1 | 5576446 | 5576450 | 4 | |
| Deletion | Chr10 | 5594132 | 5594133 | 1 | LOC_Os10g10180 |
| Deletion | Chr4 | 7418764 | 7418769 | 5 | LOC_Os04g13350 |
| Deletion | Chr8 | 8608292 | 8608293 | 1 | |
| Deletion | Chr4 | 8821122 | 8821129 | 7 | |
| Deletion | Chr1 | 9495001 | 9509000 | 14000 | 2 |
| Deletion | Chr6 | 10469935 | 10469947 | 12 | LOC_Os06g18000 |
| Deletion | Chr9 | 11677467 | 11677471 | 4 | |
| Deletion | Chr10 | 12457409 | 12457443 | 34 | |
| Deletion | Chr11 | 12580654 | 12583187 | 2533 | 2 |
| Deletion | Chr12 | 13765436 | 13765994 | 558 | |
| Deletion | Chr3 | 17230349 | 17230489 | 140 | |
| Deletion | Chr7 | 17874661 | 17874672 | 11 | LOC_Os07g30240 |
| Deletion | Chr2 | 18598981 | 18599496 | 515 | |
| Deletion | Chr7 | 18923667 | 18985895 | 62228 | 6 |
| Deletion | Chr6 | 19658166 | 19658177 | 11 | |
| Deletion | Chr12 | 23885066 | 23885067 | 1 | |
| Deletion | Chr7 | 29580526 | 29585040 | 4514 | 2 |
| Deletion | Chr2 | 29690408 | 29690416 | 8 | LOC_Os02g48500 |
| Deletion | Chr4 | 30085039 | 30085044 | 5 | |
| Deletion | Chr2 | 31226408 | 31226409 | 1 | |
| Deletion | Chr1 | 36661376 | 36661378 | 2 |
Insertions: 2
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr3 | 25922648 | 25922649 | 2 | |
| Insertion | Chr3 | 7909927 | 7909928 | 2 |
Inversions: 2
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr3 | 8047527 | 8186862 | LOC_Os03g14800 |
| Inversion | Chr3 | 31172315 | 31196127 |
Translocations: 2
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr12 | 13765988 | Chr10 | 14687397 | 2 |
| Translocation | Chr8 | 28326732 | Chr6 | 27323140 |
Tandem duplications: 1
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Tandem Duplication | Chr8 | 26016556 | 26017943 | 1387 |
