Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN589-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN589-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 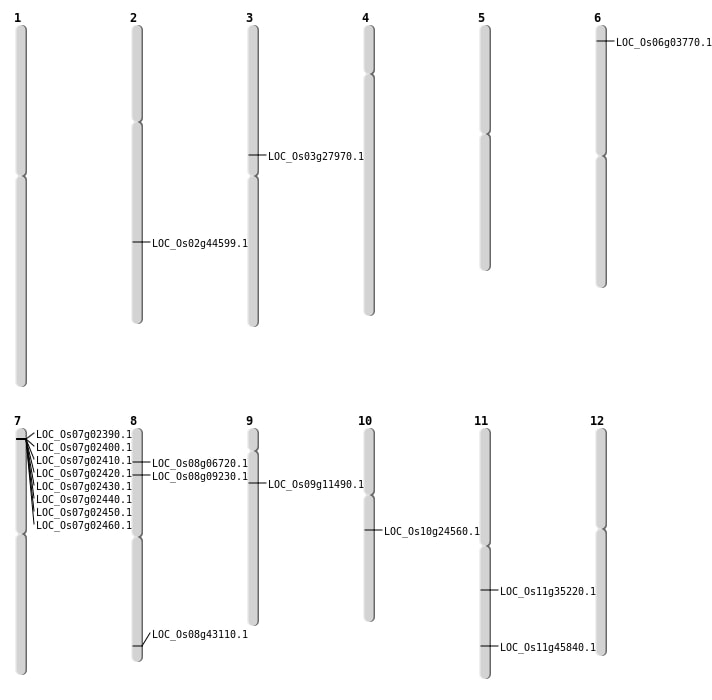
|
| Variant Information |
|---|
Single base substitutions: 33
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 11559019 | C-G | HOMO | INTERGENIC | |
| SBS | Chr1 | 17085280 | C-A | HOMO | INTRON | |
| SBS | Chr1 | 22146113 | A-G | HET | INTERGENIC | |
| SBS | Chr1 | 28884035 | T-C | HET | INTERGENIC | |
| SBS | Chr1 | 8211371 | C-T | HOMO | INTERGENIC | |
| SBS | Chr10 | 12604490 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os10g24560 |
| SBS | Chr10 | 21486934 | C-T | HOMO | UTR_3_PRIME | |
| SBS | Chr10 | 3594034 | A-G | HET | INTERGENIC | |
| SBS | Chr10 | 4015204 | T-G | HET | SYNONYMOUS_CODING | |
| SBS | Chr10 | 8269861 | G-T | HET | INTERGENIC | |
| SBS | Chr10 | 944452 | T-C | HET | INTERGENIC | |
| SBS | Chr11 | 26670244 | G-C | HET | INTERGENIC | |
| SBS | Chr11 | 3502021 | C-T | HOMO | INTERGENIC | |
| SBS | Chr12 | 22716223 | A-G | HET | INTERGENIC | |
| SBS | Chr12 | 23151762 | T-C | HET | INTRON | |
| SBS | Chr2 | 13762514 | T-C | HOMO | INTRON | |
| SBS | Chr2 | 21587693 | A-G | HET | INTERGENIC | |
| SBS | Chr3 | 28083227 | G-C | HET | INTERGENIC | |
| SBS | Chr4 | 14742644 | C-A | HET | UTR_3_PRIME | |
| SBS | Chr4 | 1784365 | C-T | HOMO | INTERGENIC | |
| SBS | Chr4 | 22922346 | C-T | HET | INTERGENIC | |
| SBS | Chr4 | 35162387 | T-C | HET | INTERGENIC | |
| SBS | Chr5 | 24587828 | C-T | HET | SYNONYMOUS_CODING | |
| SBS | Chr5 | 5357670 | A-T | HET | INTERGENIC | |
| SBS | Chr6 | 16879651 | T-A | HET | UTR_3_PRIME | |
| SBS | Chr6 | 16879652 | C-A | HET | UTR_3_PRIME | |
| SBS | Chr6 | 2033709 | G-T | HET | INTRON | |
| SBS | Chr6 | 994243 | T-C | HOMO | INTERGENIC | |
| SBS | Chr7 | 13625539 | A-T | HET | INTERGENIC | |
| SBS | Chr7 | 4528240 | G-A | HET | INTERGENIC | |
| SBS | Chr8 | 27259393 | T-A | HOMO | NON_SYNONYMOUS_CODING | LOC_Os08g43110 |
| SBS | Chr8 | 27259394 | T-A | HOMO | NON_SYNONYMOUS_CODING | LOC_Os08g43110 |
| SBS | Chr9 | 6410167 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os09g11490 |
Deletions: 22
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr7 | 819001 | 851000 | 32000 | 8 |
| Deletion | Chr6 | 994136 | 994199 | 63 | |
| Deletion | Chr6 | 1494278 | 1494287 | 9 | LOC_Os06g03770 |
| Deletion | Chr6 | 1639512 | 1639513 | 1 | |
| Deletion | Chr10 | 2815406 | 2815408 | 2 | |
| Deletion | Chr11 | 3177607 | 3177613 | 6 | |
| Deletion | Chr2 | 8212420 | 8212429 | 9 | |
| Deletion | Chr1 | 8559639 | 8559642 | 3 | |
| Deletion | Chr11 | 9678919 | 9678922 | 3 | |
| Deletion | Chr4 | 13089479 | 13089485 | 6 | |
| Deletion | Chr3 | 14257134 | 14257139 | 5 | |
| Deletion | Chr6 | 14785155 | 14785156 | 1 | |
| Deletion | Chr8 | 15179329 | 15179330 | 1 | |
| Deletion | Chr3 | 16066112 | 16066118 | 6 | LOC_Os03g27970 |
| Deletion | Chr5 | 16828624 | 16828633 | 9 | |
| Deletion | Chr2 | 19230780 | 19230781 | 1 | |
| Deletion | Chr5 | 19303553 | 19303567 | 14 | |
| Deletion | Chr11 | 20654038 | 20654043 | 5 | LOC_Os11g35220 |
| Deletion | Chr7 | 24243876 | 24243895 | 19 | |
| Deletion | Chr12 | 25218485 | 25218491 | 6 | |
| Deletion | Chr7 | 26216420 | 26216421 | 1 | |
| Deletion | Chr8 | 26252862 | 26252871 | 9 |
Insertions: 6
Inversions: 2
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr8 | 3768392 | 5372978 | LOC_Os08g06720 |
| Inversion | Chr8 | 5078171 | 5359726 | LOC_Os08g09230 |
Translocations: 2
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr12 | 17329317 | Chr11 | 27735911 | LOC_Os11g45840 |
| Translocation | Chr12 | 17329320 | Chr2 | 27027149 | LOC_Os02g44599 |
