Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN598-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN598-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 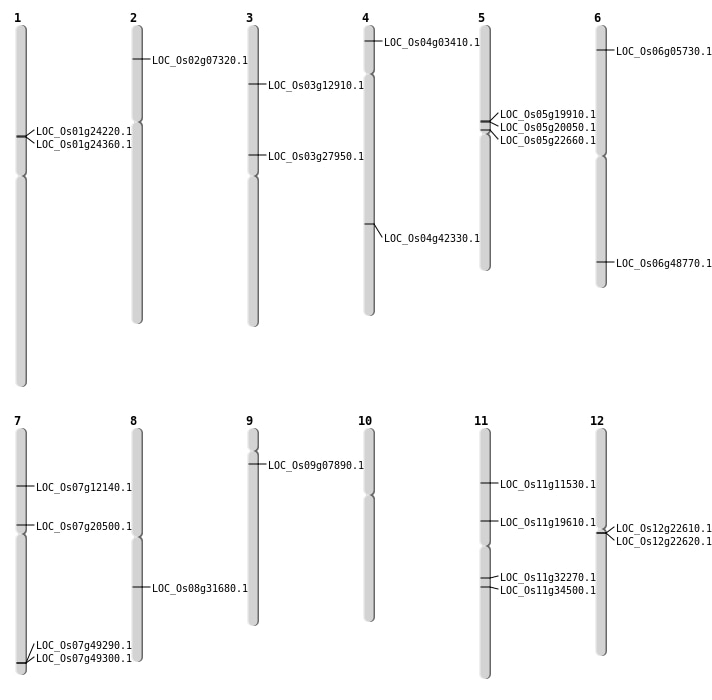
|
| Variant Information |
|---|
Single base substitutions: 39
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 13649638 | T-C | HET | NON_SYNONYMOUS_CODING | LOC_Os01g24220 |
| SBS | Chr1 | 28852334 | G-C | HOMO | INTERGENIC | |
| SBS | Chr1 | 5982728 | C-T | HET | INTERGENIC | |
| SBS | Chr1 | 8530731 | A-G | HET | INTERGENIC | |
| SBS | Chr10 | 11080900 | T-A | HOMO | INTERGENIC | |
| SBS | Chr10 | 8953453 | G-A | HET | INTERGENIC | |
| SBS | Chr11 | 10244604 | T-C | HET | INTRON | |
| SBS | Chr11 | 20210229 | C-G | HOMO | NON_SYNONYMOUS_CODING | LOC_Os11g34500 |
| SBS | Chr11 | 3696819 | C-T | HOMO | INTERGENIC | |
| SBS | Chr11 | 6132354 | G-A | HOMO | INTERGENIC | |
| SBS | Chr12 | 13311519 | T-A | HET | SPLICE_SITE_REGION | |
| SBS | Chr12 | 14185483 | C-G | HET | INTERGENIC | |
| SBS | Chr12 | 25802821 | G-A | HET | INTRON | |
| SBS | Chr2 | 12048932 | C-G | HOMO | INTRON | |
| SBS | Chr2 | 222955 | A-G | HOMO | INTERGENIC | |
| SBS | Chr2 | 2426465 | A-C | HET | INTERGENIC | |
| SBS | Chr2 | 25041584 | A-T | HET | INTERGENIC | |
| SBS | Chr3 | 16057005 | A-T | HET | NON_SYNONYMOUS_CODING | LOC_Os03g27950 |
| SBS | Chr3 | 17664451 | A-C | HET | INTRON | |
| SBS | Chr3 | 18817533 | G-A | HET | INTERGENIC | |
| SBS | Chr3 | 27228055 | A-G | HET | INTERGENIC | |
| SBS | Chr3 | 29434408 | T-C | HOMO | INTERGENIC | |
| SBS | Chr4 | 1462452 | C-T | HOMO | NON_SYNONYMOUS_CODING | LOC_Os04g03410 |
| SBS | Chr4 | 8963555 | G-A | HOMO | INTERGENIC | |
| SBS | Chr4 | 9291028 | T-C | HOMO | INTERGENIC | |
| SBS | Chr4 | 9537826 | C-T | HOMO | SYNONYMOUS_CODING | |
| SBS | Chr5 | 16873906 | C-T | HET | INTERGENIC | |
| SBS | Chr6 | 11103255 | C-T | HOMO | INTRON | |
| SBS | Chr6 | 16463973 | G-A | HOMO | INTERGENIC | |
| SBS | Chr6 | 5314768 | C-G | HET | INTERGENIC | |
| SBS | Chr6 | 8577919 | A-G | HET | INTERGENIC | |
| SBS | Chr7 | 10305221 | C-T | HET | INTERGENIC | |
| SBS | Chr7 | 21277335 | T-A | HET | INTERGENIC | |
| SBS | Chr7 | 29522502 | G-C | HOMO | NON_SYNONYMOUS_CODING | LOC_Os07g49300 |
| SBS | Chr7 | 5490354 | C-T | HOMO | INTERGENIC | |
| SBS | Chr8 | 13122882 | A-T | HET | INTERGENIC | |
| SBS | Chr8 | 14395283 | C-G | HET | INTERGENIC | |
| SBS | Chr8 | 19636259 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os08g31680 |
| SBS | Chr8 | 2596687 | C-T | HOMO | INTERGENIC |
Deletions: 25
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr2 | 3762392 | 3767572 | 5180 | LOC_Os02g07320 |
| Deletion | Chr9 | 4012982 | 4012992 | 10 | LOC_Os09g07890 |
| Deletion | Chr12 | 5929718 | 5929720 | 2 | |
| Deletion | Chr3 | 6185187 | 6185192 | 5 | |
| Deletion | Chr11 | 6411854 | 6411862 | 8 | LOC_Os11g11530 |
| Deletion | Chr7 | 6794976 | 6795001 | 25 | LOC_Os07g12140 |
| Deletion | Chr2 | 8251682 | 8251692 | 10 | |
| Deletion | Chr6 | 9174452 | 9174453 | 1 | |
| Deletion | Chr4 | 9293259 | 9293260 | 1 | |
| Deletion | Chr7 | 10226938 | 10226939 | 1 | |
| Deletion | Chr3 | 10253028 | 10253036 | 8 | |
| Deletion | Chr2 | 11864123 | 11864130 | 7 | |
| Deletion | Chr12 | 12778001 | 12786296 | 8295 | 2 |
| Deletion | Chr5 | 12860963 | 12860964 | 1 | LOC_Os05g22660 |
| Deletion | Chr4 | 13167725 | 13167728 | 3 | |
| Deletion | Chr1 | 13732153 | 13732174 | 21 | LOC_Os01g24360 |
| Deletion | Chr10 | 18755055 | 18755056 | 1 | |
| Deletion | Chr11 | 19069264 | 19069266 | 2 | LOC_Os11g32270 |
| Deletion | Chr2 | 21891047 | 21891059 | 12 | |
| Deletion | Chr2 | 25036297 | 25036299 | 2 | |
| Deletion | Chr4 | 25053772 | 25053817 | 45 | LOC_Os04g42330 |
| Deletion | Chr1 | 29163060 | 29163070 | 10 | |
| Deletion | Chr3 | 29434458 | 29434459 | 1 | |
| Deletion | Chr7 | 29522111 | 29522126 | 15 | LOC_Os07g49290 |
| Deletion | Chr4 | 34611621 | 34611622 | 1 |
Insertions: 5
Inversions: 7
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr6 | 2588642 | 2938739 | LOC_Os06g05730 |
| Inversion | Chr7 | 10649232 | 11836711 | 2 |
| Inversion | Chr7 | 10649395 | 11836714 | LOC_Os07g20500 |
| Inversion | Chr5 | 11613287 | 11723507 | 2 |
| Inversion | Chr12 | 14185275 | 16314354 | |
| Inversion | Chr6 | 28224586 | 29521101 | LOC_Os06g48770 |
| Inversion | Chr6 | 28224606 | 29521099 | LOC_Os06g48770 |
Translocations: 6
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr3 | 6959011 | Chr2 | 3762755 | 2 |
| Translocation | Chr3 | 6959021 | Chr2 | 3762397 | 2 |
| Translocation | Chr7 | 10649232 | Chr5 | 11479649 | 2 |
| Translocation | Chr7 | 10649393 | Chr5 | 11613401 | LOC_Os05g19910 |
| Translocation | Chr11 | 11303517 | Chr6 | 3063650 | LOC_Os11g19610 |
| Translocation | Chr11 | 11303525 | Chr6 | 3065976 | LOC_Os11g19610 |
