Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN621-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN621-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 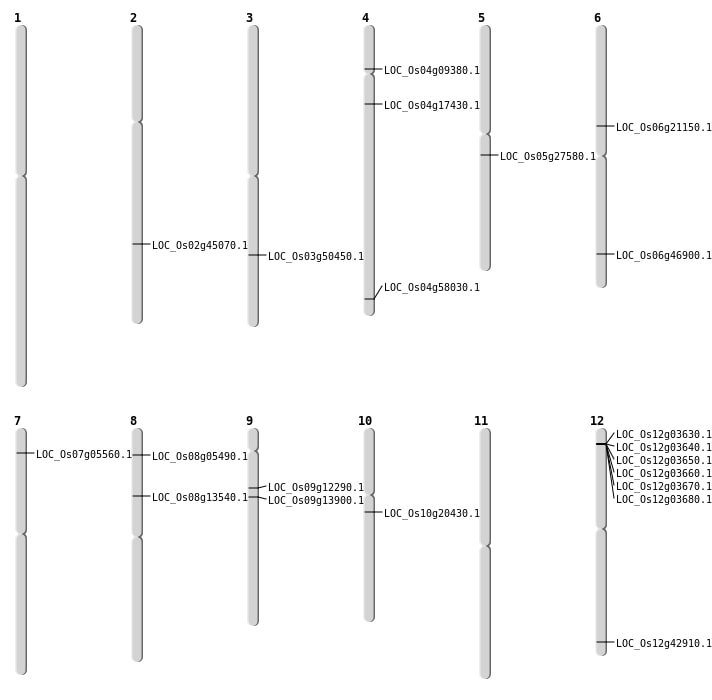
|
| Variant Information |
|---|
Single base substitutions: 39
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 2023137 | C-T | HET | INTERGENIC | |
| SBS | Chr10 | 12578385 | C-T | HET | INTERGENIC | |
| SBS | Chr11 | 18303786 | G-T | HOMO | INTERGENIC | |
| SBS | Chr11 | 2635674 | C-A | HOMO | INTERGENIC | |
| SBS | Chr11 | 27141253 | C-T | HOMO | INTERGENIC | |
| SBS | Chr12 | 11519557 | A-C | HOMO | INTERGENIC | |
| SBS | Chr12 | 1608993 | A-T | HOMO | INTRON | |
| SBS | Chr12 | 26204952 | T-A | HET | INTERGENIC | |
| SBS | Chr12 | 26204953 | G-C | HET | INTERGENIC | |
| SBS | Chr2 | 15403476 | A-T | HET | INTERGENIC | |
| SBS | Chr2 | 16015660 | G-A | HET | INTERGENIC | |
| SBS | Chr2 | 21199712 | T-A | HET | INTERGENIC | |
| SBS | Chr2 | 27332593 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os02g45070 |
| SBS | Chr3 | 1982270 | T-A | HET | INTERGENIC | |
| SBS | Chr3 | 23142619 | C-T | HET | INTERGENIC | |
| SBS | Chr4 | 11220265 | G-T | HOMO | SYNONYMOUS_CODING | |
| SBS | Chr4 | 11768602 | G-A | HOMO | INTRON | |
| SBS | Chr4 | 11768603 | T-A | HOMO | INTRON | |
| SBS | Chr4 | 19188504 | A-G | HET | INTERGENIC | |
| SBS | Chr4 | 6604567 | G-A | HET | SYNONYMOUS_CODING | |
| SBS | Chr5 | 1196924 | T-C | HOMO | INTRON | |
| SBS | Chr5 | 12992729 | C-A | HET | INTERGENIC | |
| SBS | Chr5 | 14717094 | C-T | HET | INTRON | |
| SBS | Chr5 | 23888499 | C-A | HET | INTRON | |
| SBS | Chr5 | 27777300 | A-T | HET | INTERGENIC | |
| SBS | Chr5 | 8654875 | C-T | HOMO | INTERGENIC | |
| SBS | Chr6 | 11450109 | G-A | HOMO | UTR_3_PRIME | |
| SBS | Chr6 | 5888268 | A-T | HET | INTERGENIC | |
| SBS | Chr7 | 2599614 | C-A | HET | NON_SYNONYMOUS_CODING | LOC_Os07g05560 |
| SBS | Chr8 | 12790588 | C-T | HOMO | INTRON | |
| SBS | Chr8 | 14075713 | G-A | HOMO | INTERGENIC | |
| SBS | Chr8 | 20473554 | G-A | HET | INTERGENIC | |
| SBS | Chr8 | 25959797 | A-C | HET | INTRON | |
| SBS | Chr8 | 5015928 | C-A | HET | INTRON | |
| SBS | Chr8 | 8049933 | C-T | HOMO | NON_SYNONYMOUS_CODING | LOC_Os08g13540 |
| SBS | Chr9 | 16377140 | G-A | HOMO | SYNONYMOUS_CODING | |
| SBS | Chr9 | 18976919 | C-T | HET | INTERGENIC | |
| SBS | Chr9 | 7004595 | G-T | HET | NON_SYNONYMOUS_CODING | LOC_Os09g12290 |
| SBS | Chr9 | 8184236 | G-A | HET | STOP_GAINED | LOC_Os09g13900 |
Deletions: 25
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr9 | 1194875 | 1194877 | 2 | |
| Deletion | Chr12 | 1454597 | 1487814 | 33217 | 6 |
| Deletion | Chr6 | 4210914 | 4210915 | 1 | |
| Deletion | Chr9 | 8920629 | 8920642 | 13 | |
| Deletion | Chr4 | 9537413 | 9537418 | 5 | LOC_Os04g17430 |
| Deletion | Chr3 | 10153325 | 10153330 | 5 | |
| Deletion | Chr10 | 10275102 | 10275189 | 87 | LOC_Os10g20430 |
| Deletion | Chr2 | 15206312 | 15206321 | 9 | |
| Deletion | Chr7 | 15597847 | 15597852 | 5 | |
| Deletion | Chr9 | 16377366 | 16377368 | 2 | |
| Deletion | Chr9 | 16421608 | 16421632 | 24 | |
| Deletion | Chr8 | 18956475 | 18956476 | 1 | |
| Deletion | Chr12 | 19952656 | 19952658 | 2 | |
| Deletion | Chr2 | 20555185 | 20555188 | 3 | |
| Deletion | Chr5 | 20588656 | 20588691 | 35 | |
| Deletion | Chr5 | 20627818 | 20627819 | 1 | |
| Deletion | Chr11 | 25298896 | 25306952 | 8056 | |
| Deletion | Chr12 | 26669655 | 26669700 | 45 | LOC_Os12g42910 |
| Deletion | Chr6 | 28463974 | 28463975 | 1 | LOC_Os06g46900 |
| Deletion | Chr7 | 28500430 | 28500431 | 1 | |
| Deletion | Chr3 | 28798486 | 28798489 | 3 | LOC_Os03g50450 |
| Deletion | Chr3 | 28798492 | 28798495 | 3 | LOC_Os03g50450 |
| Deletion | Chr2 | 33626526 | 33626528 | 2 | |
| Deletion | Chr4 | 34564813 | 34564814 | 1 | LOC_Os04g58030 |
| Deletion | Chr4 | 35247154 | 35247161 | 7 |
Insertions: 3
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr1 | 23047852 | 23047853 | 2 | |
| Insertion | Chr6 | 12217923 | 12217923 | 1 | LOC_Os06g21150 |
| Insertion | Chr6 | 28300445 | 28300448 | 4 |
No Inversion
Translocations: 5
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr9 | 8523391 | Chr6 | 21524080 | |
| Translocation | Chr9 | 10809754 | Chr8 | 2926364 | LOC_Os08g05490 |
| Translocation | Chr5 | 16047420 | Chr4 | 11221941 | LOC_Os05g27580 |
| Translocation | Chr5 | 16047672 | Chr4 | 11221934 | LOC_Os05g27580 |
| Translocation | Chr7 | 17250530 | Chr4 | 5000290 | LOC_Os04g09380 |
